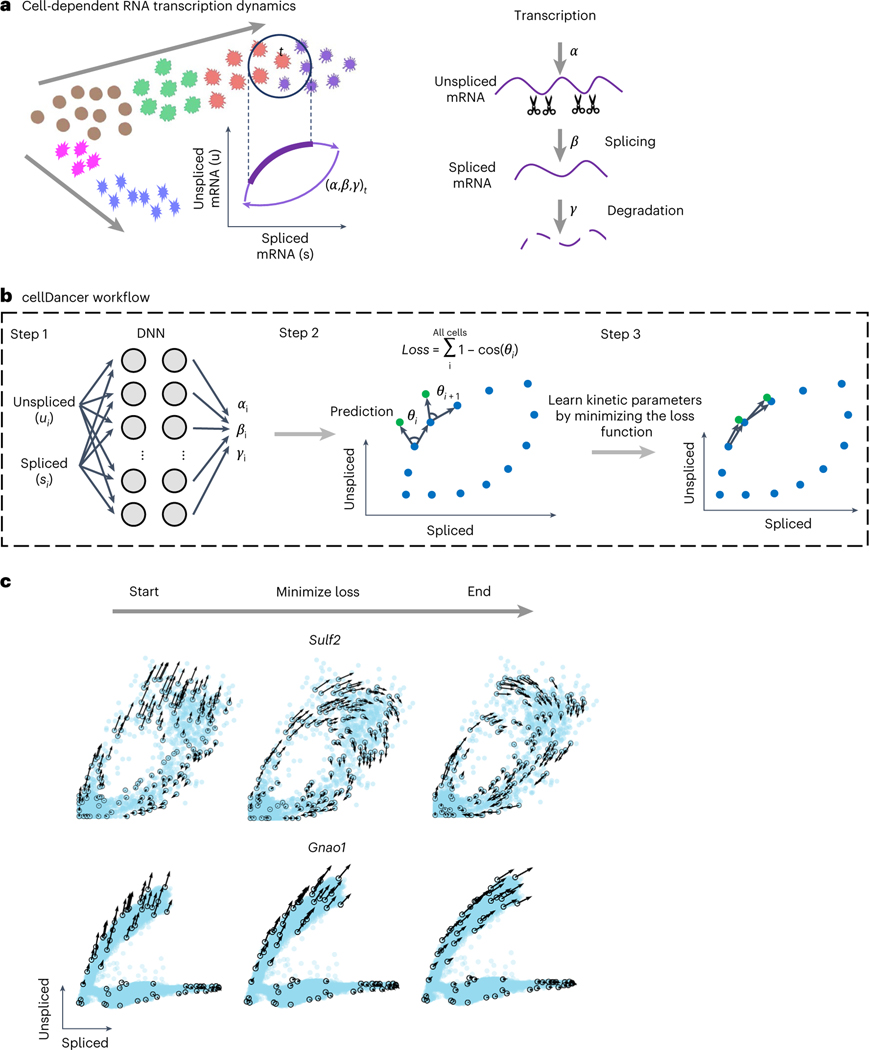
Fig. 1 |. Predicting RNA velocity in localized cell populations via DNNs.
a, Transcription dynamics of the premature (unspliced) and mature (spliced) mRNAs are governed by the transcription (), splicing () and degradation () rates. Multi-kinetics genes involve multiple-lineage and/or multi-stage transitions of the cellular states; hence, cell-dependent rates (, , )t are required to accurately capture the transcription dynamics of those genes. In the illustration, the (, , )t for cell t are computed by locating the future state cell in the neighboring cells of t (‘local environment’), assuming that the cells in the local environment share the same (, , ). b, cellDancer uses a DNN to predict cell-specific , and for each gene. The DNN consists of an input layer with the spliced and unspliced mRNA abundances , two fully connected hidden layers each with 100 nodes and an output layer yielding cell-specific , and . The loss function is defined as the sum of every cell’s cosine similarity of predicted and observed velocity vectors. The DNN is iteratively optimized by minimizing the loss function. c, The progress of minimizing the loss function. RNA velocities for the examples of the mono-kinetic gene Sulf2 in pancreatic endocrinogenesis, and the multi-lineage gene Gnao1 in mouse hippocampus maturation is projected onto the phase portraits during the training process of their DNNs.