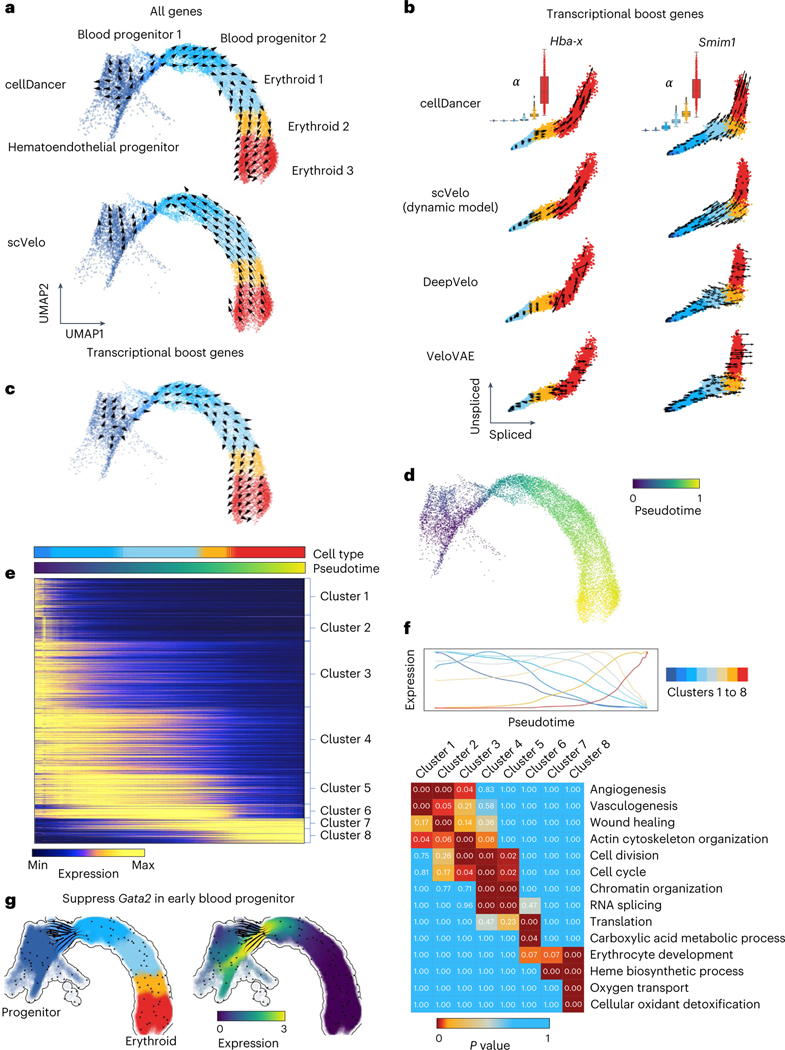
Fig. 2 |. Delineating gastrulation erythroid maturation and resolving transcriptional boost.
a, The velocities derived from cellDancer (top) are consistent with the erythroid differentiation but opposite in scVelo dynamic model (bottom) by using all genes. b, The velocities derived from cellDancer, scVelo dynamic model, DeepVelo and VeloVAE for the transcriptional boost genes (Hba-x and Smim1) are illustrated on the phase portraits. The cells are colored according to the cell types. The box plots of for each cell type predicted by cellDancer are included to show the boost in the rates in the course of erythroid maturation, especially in erythroid 3. c, The velocities derived from cellDancer for gastrulation erythroid maturation using transcriptional boost genes are projected on the UMAP of the original work, demonstrating that cellDancer can infer the correct cell differentiation direction by using only the transcriptional boost genes. d, Gene-shared pseudotime on UMAP is consistent with the progression of gastrulation erythroid maturation. e, Genes that show high similarity in transcriptional changes along time are classified into eight clusters according to their transcriptional changes. The heat map describes the expression of the genes along time (rows: genes; columns: cells ordered according to the pseudotime). Genes were selected by Pearson correlation coefficient (R2) > 0.8. f, Average expression of each cluster along the pseudotime (top) and the enriched pathways for each cluster of genes (bottom) (Benjamini–Hochberg procedure, one-sided, P < 0.05). P value indicates the significance of enrichment of a pathway in Fisher’s exact test. g, In silico perturbation analysis by dynamo shows a critical role of Gata2 in hematopoiesis.