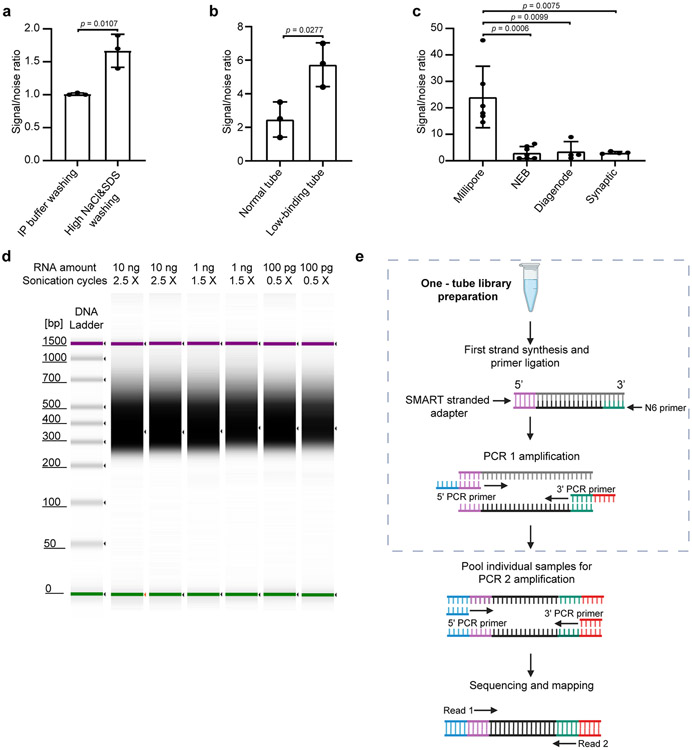
Extended Data Fig. 1. ∣. Optimization and development of picoMeRIP-seq.
a, qPCR assessment of signal-to-noise ratio for the evaluation of the effect of increased stringency of the wash buffers. Mouse liver polyA selected RNA (10 ng) was used in these experiments. The previously validated m6A positive (Pdzd8) and negative (Rdh10) loci were used to calculate the signal-to-noise ratio (See Methods). Data are presented as mean ± standard deviation (SD), with n = 3 independent experiments for each experimental condition. Unpaired, two-tailed t-test was used. b, qPCR assessment of signal-to-noise ratio for the evaluation of the effect of low-binding tubes. Mouse liver polyA selected RNA (10 ng) was used in these experiments. Data are presented as mean ± SD values, with n = 3 independent experiments for each experimental condition. Unpaired, two-tailed t-test was used. c, qPCR assessment of signal-to-noise ratio for the optimized picoMeRIP method with different antibodies. Mouse liver polyA selected RNA (10 ng) was used in these experiments. Data are presented as mean ± SD values, with n = 6; 7; 4; 4 independent experiments for Millipore, NEB, Diagenode, Synaptic Systems antibodies, respectively. Unpaired, two-tailed t-test was used. d, Synthetic gel image from an Agilent 2100 Bioanalyzer electrophoreses run showing the size of sequencing libraries generated from picoMeRIP-seq of 10 ng, 1 ng and 100 pg mouse liver polyA selected RNA. Library preparation adds 139 bp to the size of the immunoprecipitated RNA fragments. The top line indicates the applied number of sonication cycles with the Hielscher UP100H sonicator. Each cycle is 30 seconds sonication plus 30 seconds on ice. e, Tailored SMART library preparation with a single-tube protocol to include as much as possible of the immunoprecipitated RNA. The SMART scN6 Primer (green) random primers allows the generation of cDNA from all immunoprecipitated RNA fragments. After the reverse transcription, the SMART Stranded Adapter (pink) will be linked with the cDNA product. Next, a first round of PCR amplification (PCR 1) adds full-length Illumina adapters, including barcodes. To minimize the number of samples to be processed downstream, individual samples can be pooled together for a second round of PCR amplification (PCR 2) using primers universal to all libraries. The final library is compatible with any Illumina sequencing platform. Created with BioRender.com.