Fig. 1.
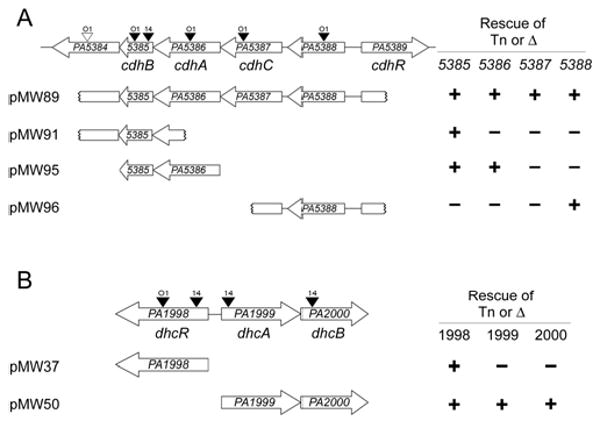
Two regions of the P. aeruginosa genome contain genes required for carnitine catabolism to glycine betaine. A) The PA5384 to PA5389 region contains genes involved in the first step of carnitine catabolism, conversion to 3-dhc. B) The PA1998 to PA2000 region contains genes predicted to be involved in conversion of 3-dhc to glycine betaine. Transposon insertions are marked with triangles above the corresponding gene with a labeled to show the strain carrying the transposon; O1 = PAO1, 14 = PA14. Black triangles represent transposon insertions that could not grown on carnitine and white triangles represent insertions that did not alter growth on carnitine. Complementation constructs are diagramed below each genomic region with notation at the left denoting capability (+) or failure (-) to rescue a deletion or transposon insertion into the gene at the top of the column. Growth rates for strains summarized here can be seen in Table S2. The gene names below each genomic region are based on the naming scheme proposed in this report.
