Fig. 2 |. High sensitivity of USeqFISH detects short mutations and barcodes in the AAV genome in vitro and in vivo.
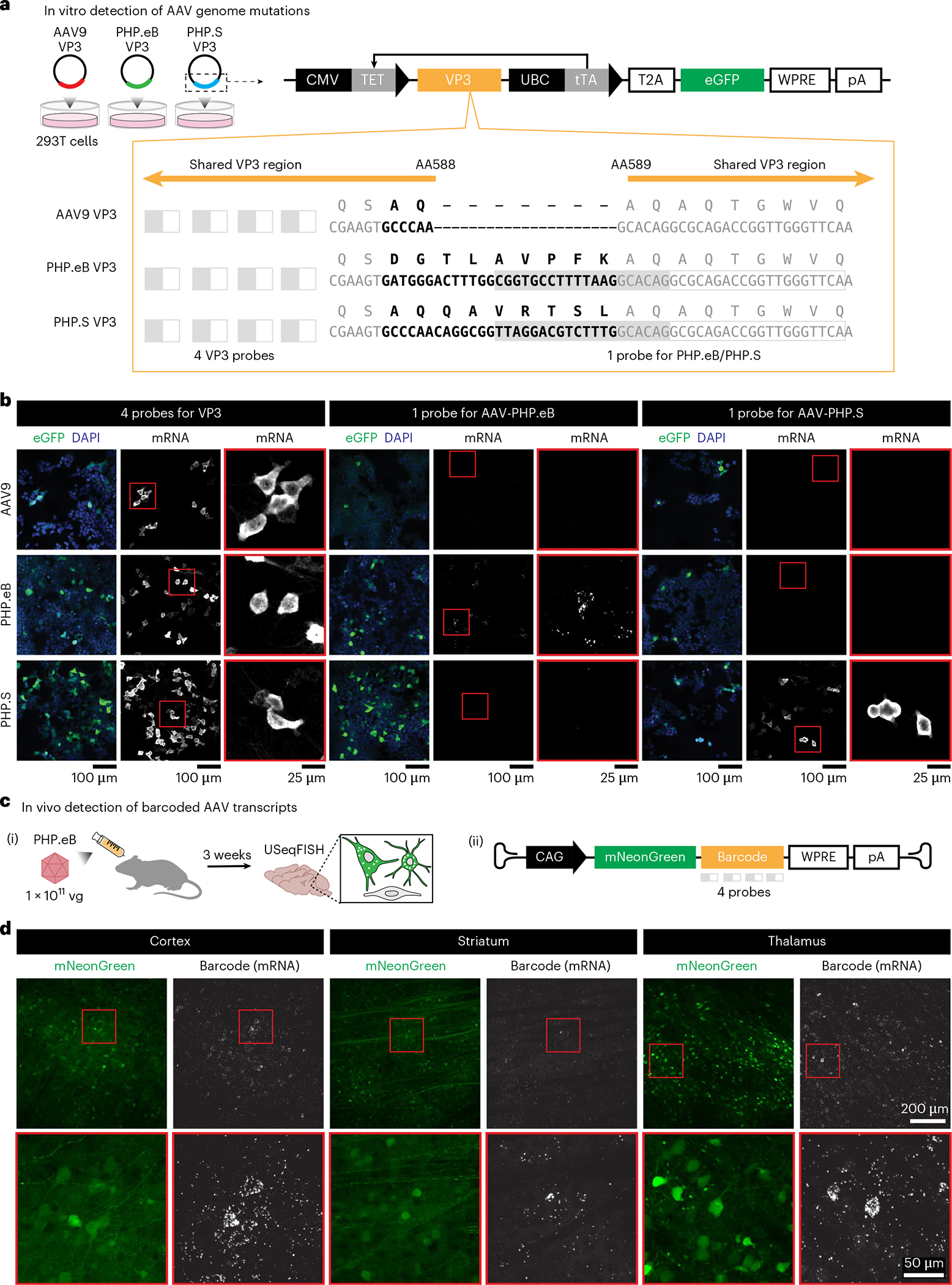
a, Three plasmids were designed to carry the VP3 of AAV9, AAV-PHP.eB (‘PHP.eB’) and AAV-PHP.S (‘PHP.S’) with eGFP. AAV-PHP.eB and AAV-PHP.S have distinct 9-AA and 7-AA mutations (bold letters) in the same location (AA588) of the AAV9 VP3 sequence. After transfecting into HEK293T cells, we detected the transcripts of each plasmid using the following probes (gray filled boxes indicate the padlock target sequence, and gray outlined boxes indicate the primer target sequence): four probes against the shared VP3 sequence, one probe against the insertion of AAV-PHP.eB and one probe against the insertion of AAV-PHP.S. For the probes against each insertion, we used the same primers for AAV-PHP.eB and AAV-PHP.S but distinct padlocks that differed by 14 nt. b, Detection of the VP3 transcripts with four probes for VP3, one probe for AAV-PHP.eB and one probe for AAV-PHP.S in HEK293T cells expressing the VP3 of AAV9, AAV-PHP.eB and AAV-PHP.S. c, For in vivo detection, we designed a viral genome carrying mNeonGreen and a barcode and systemically delivered it to adult mice using AAV-PHP.eB at a dose of 1 × 1011 vg per mouse. At 3 weeks after injection, we used USeqFISH with probes against the barcode to detect viral transcripts in tissue. d, Detection of viral barcodes (‘Barcode (mRNA)’) in cells expressing mNeonGreen (green) in various mouse brain regions (cortex, striatum and thalamus).
