Fig. 6 |. USeqFISH application to NHPs: in situ AAV detection and integrative analysis of cell morphology and transcriptional profiles.
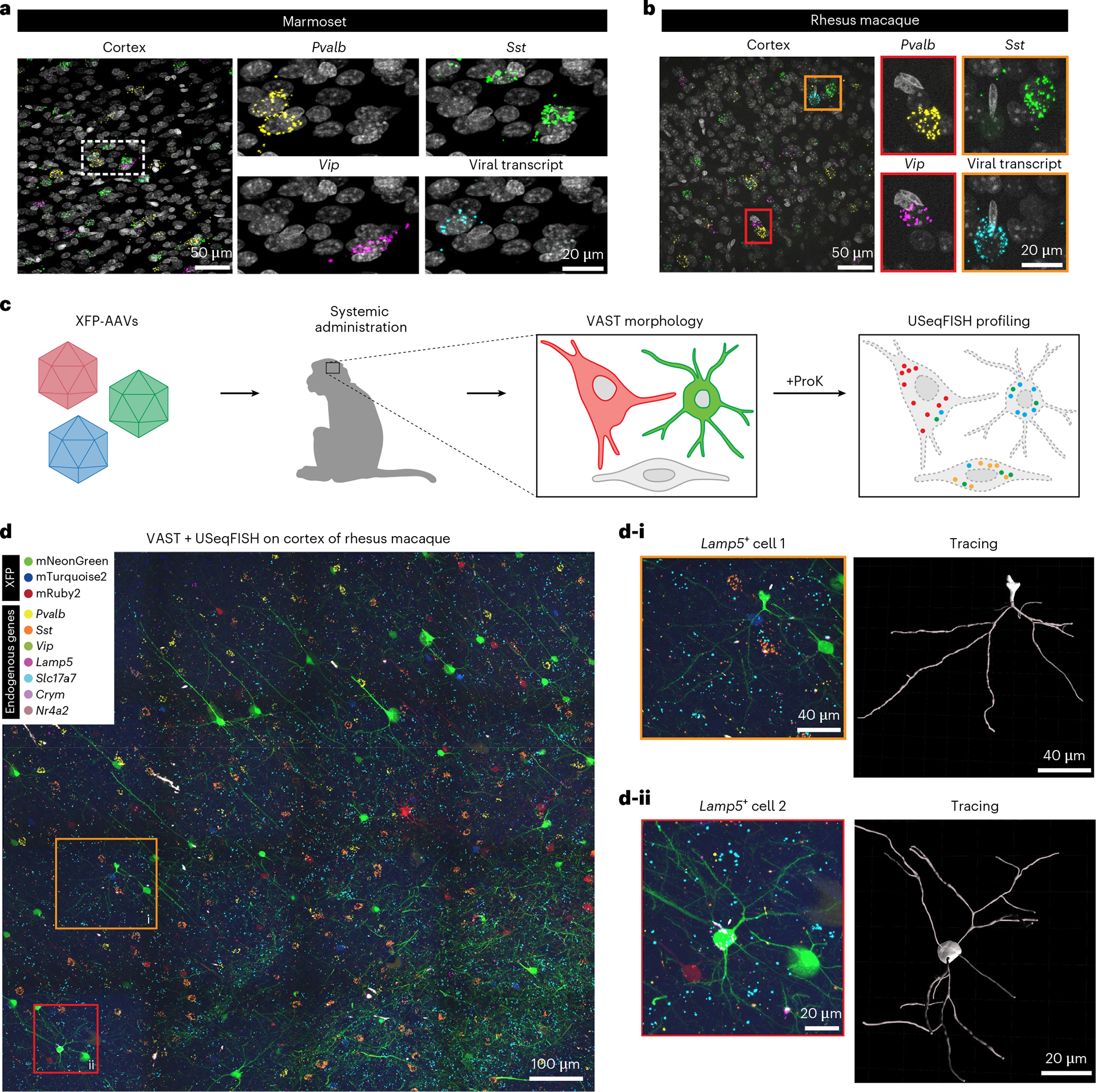
a,b, We applied USeqFISH to brain tissue slices of marmoset (a) and rhesus macaque (b) to which our viruses were administered (eight pooled variants for the marmoset and AAV.CAP-Mac for the rhesus macaque) with probes against three endogenous genes (yellow: Pvalb; green: Sst; magenta: Vip) and the coding sequence of each viral genome (human frataxin for the marmoset and mNeonGreen for the rhesus macaque (cyan); FPs were quenched by proteinase K (ProK) treatment). The representative images show that USeqFISH is applicable to these two NHP species with species-specific probes. c, Schematic of procedure of vector-assisted spectral tracing (VAST) and subsequent USeqFISH profiling of the rhesus macaque brain. We systemically delivered a cocktail of three AAV.CAP-Mac viruses packaging mNeonGreen, mTurquoise2 or mRuby2 to an infant rhesus macaque and recovered the brain. This brain exhibited a variety of colors, coming from stochastic expression of the three FPs, allowing us to trace single-cell morphologies. We additionally labeled seven endogenous genes (Pvalb, Sst, Vip, Lamp5, Slc17a7, Crym and Nr4a2) using USeqFISH in the same tissue to identify transcriptionally defined cell types and their morphology. d, Representative image of integration of VAST and USeqFISH with seven cell marker genes in the rhesus macaque brain and examples of two cells (yellow outlined box: i; red outlined box: ii) identifying both cell type and morphology.
