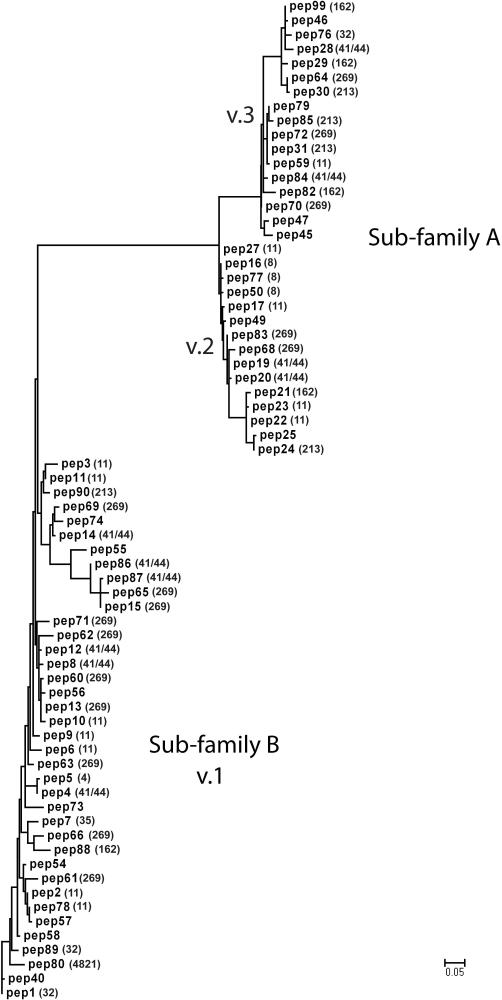
Figure 1.
Phylogram of fHbp based on 70 unique amino acid sequences. For each sequence, the peptide identification number assigned in the fHbp peptide database at http://Neisseria.org is shown and, if known, the multi-locus sequence type (MLST) clonal complex is shown in parentheses. The lower left branch shows variant group 1 as defined by Masignani et al (Masignani et al., 2003) (sub-family B of Fletcher et al (Fletcher et al., 2004)); Sub-family A contained two branches, variant groups 2 and 3. The phylogram was constructed by multiple sequence alignment as described in Methods and supplementary Table S2. The scale bar shown at the bottom indicates 5 amino acid changes per 100 residues.