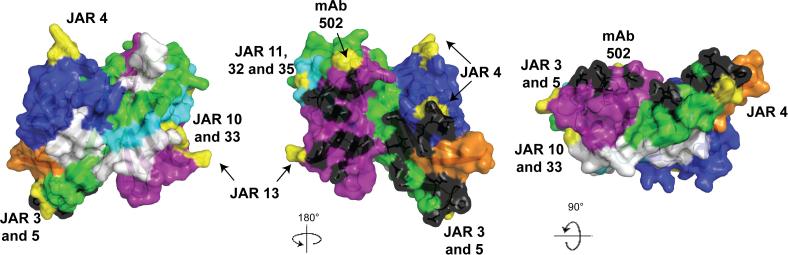
Figure 7.
Space-filled structural models of factor H binding protein based on the coordinates of fHbp in a complex with a fragment of human factor H (Schneider et al., 2009). The five variable segments VA through VE are depicted in different colors (VA, blue; VB, orange; VC, green; VD, aqua; VE, violet) and the invariant blocks of residues separating each of the variable segments are shown in white. The model in the middle was generated by rotation of 180° around the Y-axis relative to the respective model on the far left. The model on the right was generated by a 90° rotation around the X-axis relative to the model in the middle. The factor H contact residues are depicted in black, and the residues affecting the epitopes of anti-fHbp mAbs are shown in yellow. The figure was constructed with PyMol (http://www.pymol.org).