Figure 1. Mechanistic insights from epigenomic signatures of chromosomal syndromes with ASD.
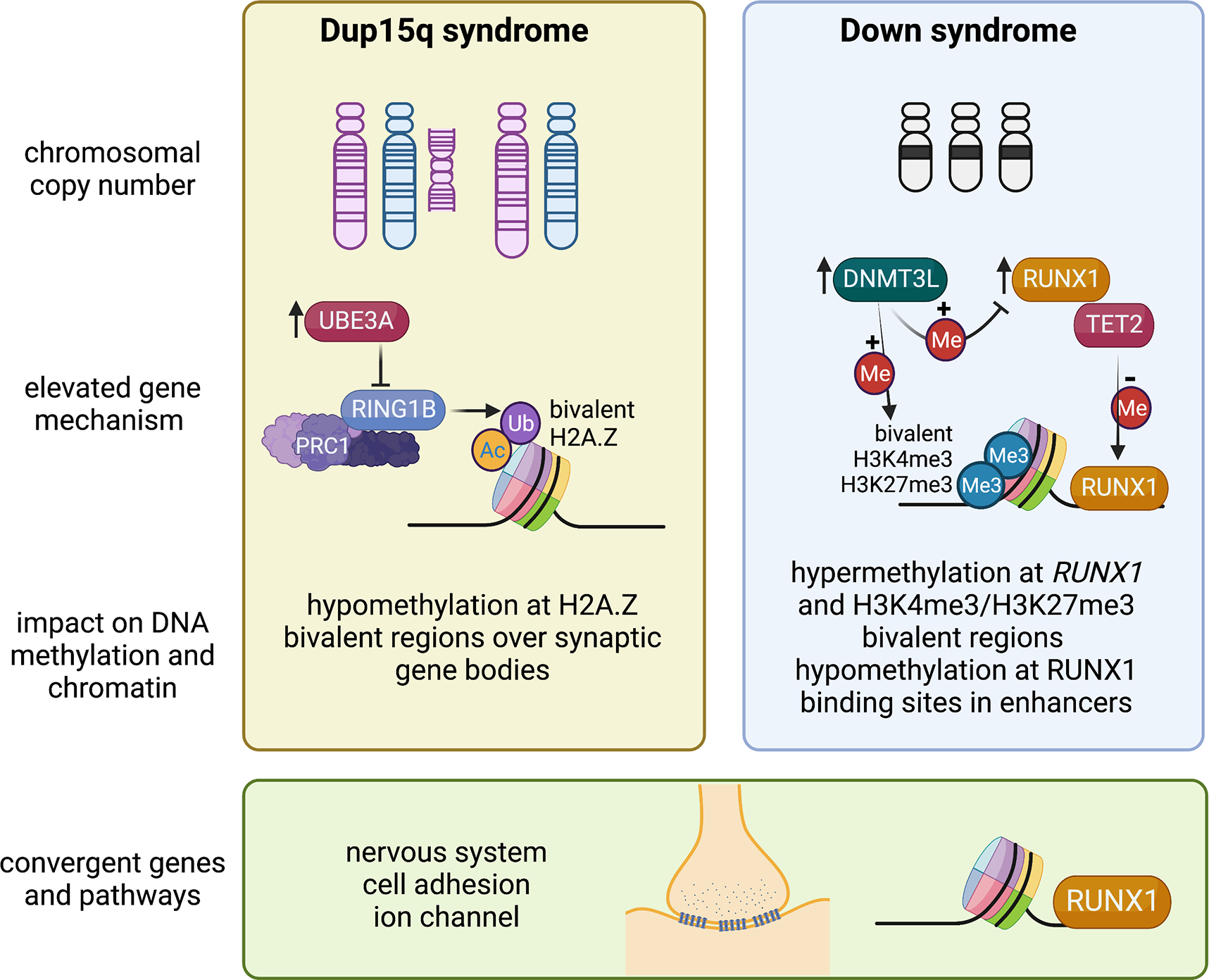
Examples are shown of two disorders caused by a large chromosomal duplication, 15q11-q13 duplication syndrome (Dup15q, left panel), or aneuploidy, Down syndrome (trisomy 21, right panel). In Dup15q syndrome, duplications are either supernumerary (left) or interstitial (right) and depend on parent of origin, as only maternal (pink chromosome) duplications are associated with ASD. Elevated levels of the imprinted gene UBE3A are predicted to initiate the pathogensis of Dup15q syndrome. UBE3A is an E3 ubiquitin ligase that targets a different E3 ubiquitin ligase called RING1B, part of the PRC1 complex, that monoubiquitinates H2A and H2A.Z, resulting in a maintenance of bivalency (acetylation and ubiquitination). The epigenomic signature of Dup15q brain and neuronal cell line models has revealed hypomethylation at H2A.Z bivalent regions over synaptic genes due to reduced RING1B levels. In contrast, Down syndrome brain is characterized by both hypermethylation and hypomethylation and two chromosome 21-encoded proteins are distinctly implicated in each process. Elevated DNMT3L increases methylation over regions of bivalent chromatin marked by H3K4me3 and H3K27me3, including the chromosome 21 locus RUNX1, encoding a developmental transcription factor. The regions hypomethylated in Down syndrome newborn blood were enriched for RUNX1 binding sites, suggesting that RUNX1 targets these sites for demethylation from its known association with TET2. While the mechanisms behind the epigenomic signatures of Dup15q and Down syndromes are distinct, synaptic gene pathways are apparent in both and the RUNX1 locus shows differential methylation in both syndromes. Created with BioRender.com.
