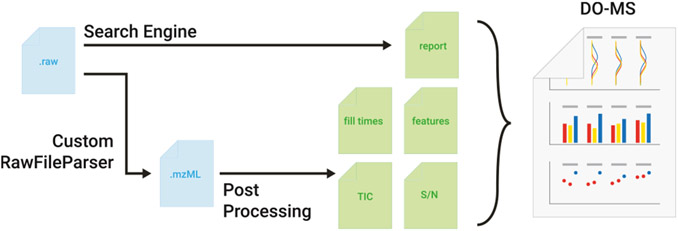
Figure 1.
Schematic of the DO-MS pipeline version 2.0. A schematic of the processing and intermediate steps of the updated DO-MS pipeline. Input files (blue) in the raw format are searched by a search engine (the default one is DIA-NN13) and converted to mzML using a custom version of the ThermoRawFile parser.41 The search report from DIA-NN and the mzML are then used by the post-processing step to analyze and display data about MS1 and MS2 accumulation times, TIC information, precursor-wise signal-to-noise levels, and MS1 features.