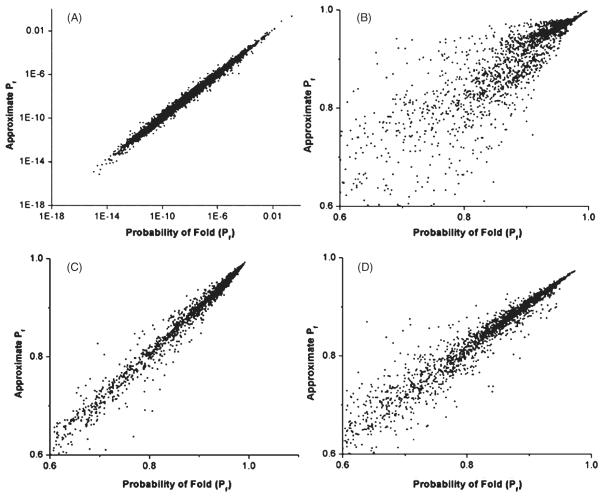
Figure 5.
Relationship between the true probability of folding to the target sequence and the approximate probability of folding estimated from a limited sample of structures. Sequences were sampled either randomly (A) or else all mutants were sampled for 800 generations of the evolutionary process subsequent to equilibrium (B–D). The approximate probability of folding was estimated from a sample of random structures plus the same number of structures closest to the native structure in terms of shared contact pairs (B). The random sample was treated as a representative of the remaining unsampled structural ensemble, and thus multiplied by the inverse of its proportional representation of this ensemble (see methods). In (C) and (D), each category of structural distance from the native or target structure was sampled separately, and the contribution of each distance category to the overall partition function was also estimated separately. The number of random and adjacent structures used in the approximations was 5,000 in all cases except (D), for which 500 distance-based random structures and 500 adjacent structures were used.