Figure 1.
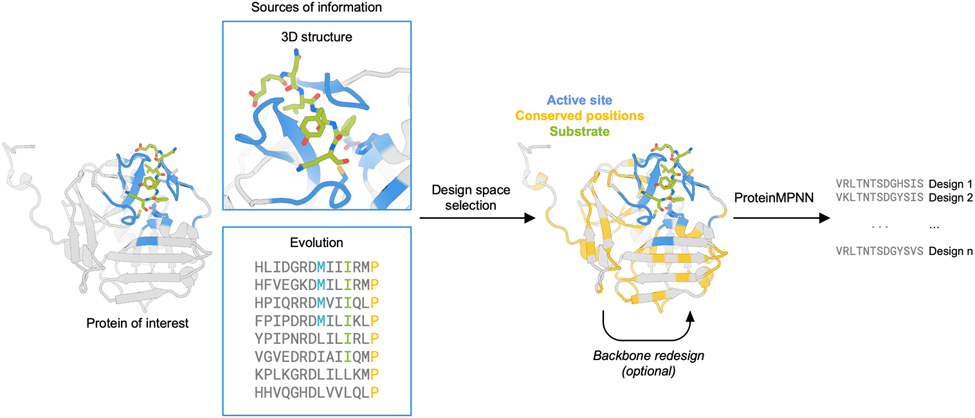
Design strategy for optimization of protein expression and stability using ProteinMPNN. The design space is chosen to preserve native protein function by fixing the amino acid identities of residues close to the ligand and those that are highly conserved in multiple sequence alignments. The protein backbone structure and fixed position information of amino acids are input into ProteinMPNN, which generates new amino acid sequences likely to fold to the input structure. The backbone structure in loop regions can optionally be remodeled using RoseTTAfold joint inpainting to further idealize the input protein.
