Figure 3.
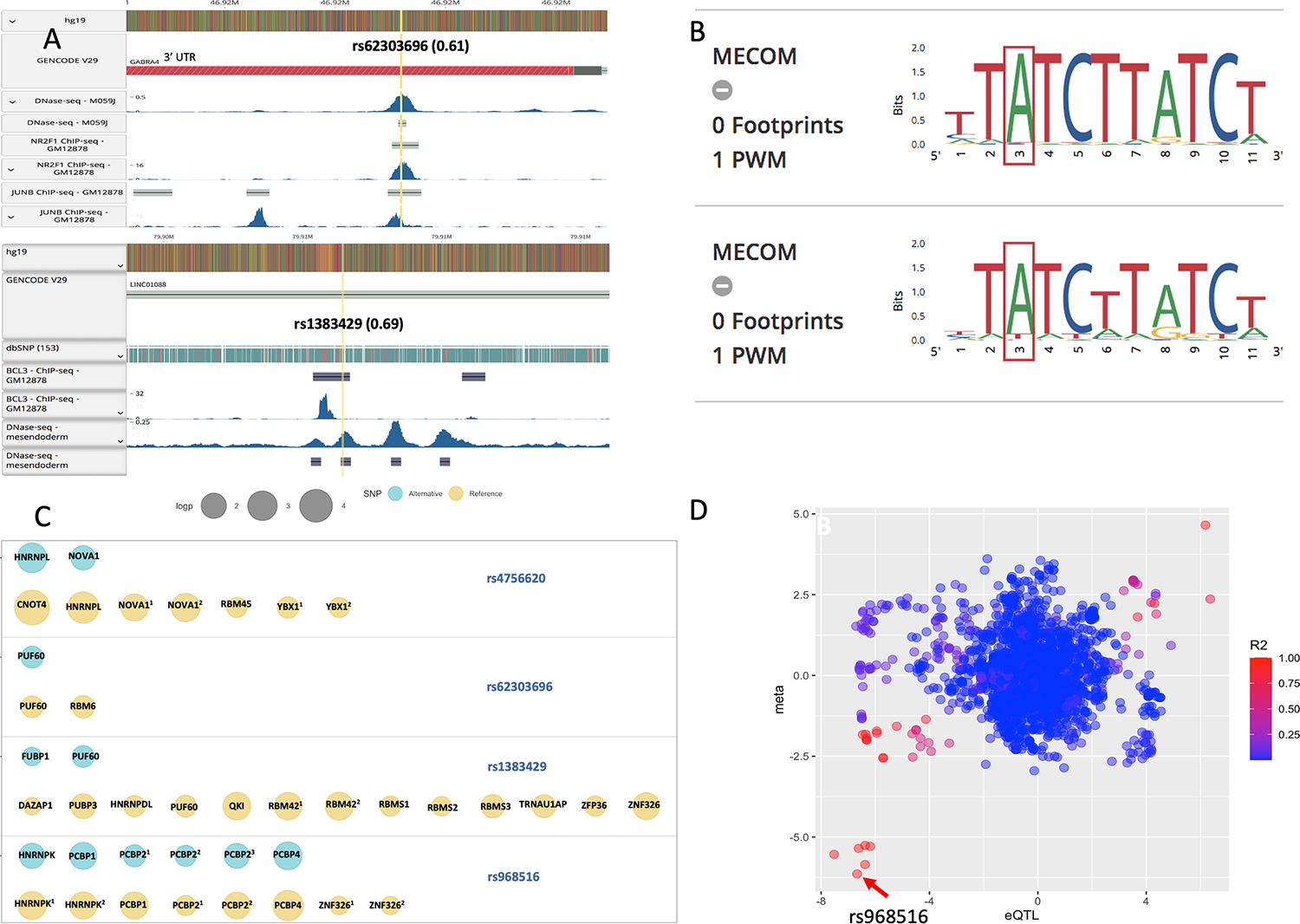
functional analysis of the novel variants identified in ever- and never-smoking lung cancer. A, CHIP-seq peaks were identified at rs62303696 and rs1383429 by query from RegulomeDB. 0.61 and 0.69 in the brackets indicated the calculated probability of being a regulatory variant. B, the predicted sequence motif including the highly conservative allele at rs1383429. C, annotation results from RBPmap. The RBPs (RNA Binding Proteins) with significant sequence motifs (P < 0.05) between reference (yellow) and alternative (green) alleles were displayed for each variant located within coding gene. The size of circle indicates the significance of the sequence motif. D, eQTL analysis for rs968516 and LCNL1 gene. The X axis denotes the Z-score from association analysis between genotype and LCNL1 gene expression from GTEx data from lung tissues. Y axis denotes the Z-score from GWAS analysis. The color intensity reflected the extent of linkage disequilibrium index (r2) with SNP rs968516 denoted by arrow.
