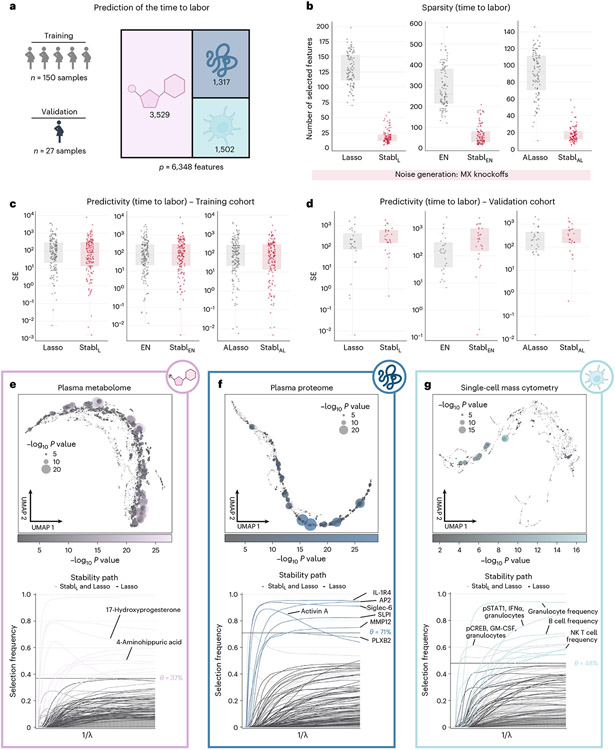
Fig. 5 ∣. Stabl’s performance on a triple-omic data integration task.
a, Clinical case study 3: prediction of the time to labor from longitudinal assessment of plasma proteomic (SomaLogic), metabolomic (untargeted mass spectrometry) and single-cell mass cytometry data in two independent cohorts of pregnant individuals. b, Sparsity performances (number of features selected across CV iterations, median and IQR) for StablL (left), StablEN (middle) and StablAL (right) compared to their respective SRM (late-fusion data integration method) across n = 100 CV iterations. c,d, Predictivity performances as squared error (SE) on the training (n = 150 samples, c) and validation (n = 27 samples, d) datasets for StablL (left), StablEN (middle) and StablAL (right). StablSRM performances are shown using MX knockoffs. Results using random permutations are shown for StablL in Supplementary Table 5. Median and IQR values comparing StablSRM performances to their cognate SRMs are listed in Supplementary Table 5. e–g, UMAP visualization (upper) and stability path (lower) of the metabolomic (e), plasma proteomic (f) and single-cell mass cytometry (g) datasets. UMAP node size and color are proportional to the strength of association with the outcome. Stability path graphs denote features selected by StablL. The data-driven reliability threshold is computed for each individual omic dataset and is indicated by a dotted line. Significance of the association with the outcome was calculated using Pearson’s correlation. Box plots indicate median and IQR; whiskers indicate 1.5× IQR.