Extended Data Fig. 6.
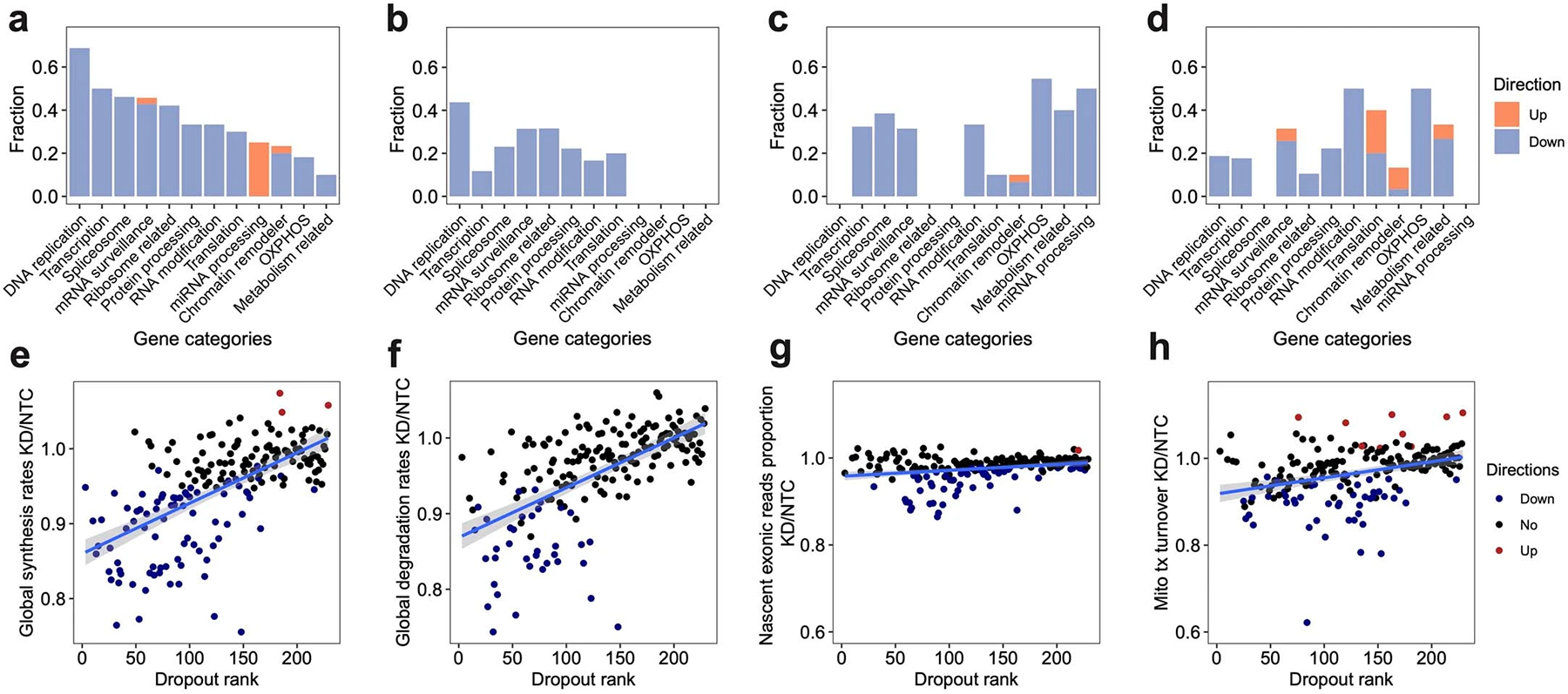
a-d. For each gene category, we calculated the fraction of genetic perturbations associated with significant changes in global synthesis rates (a), global degradation rates (b), proportions of exonic reads in the nascent transcriptome (c), and proportions of mitochondrial nascent reads (d). Overall global transcription could be affected by more genes than degradation. Perturbation on essential genes, such as DNA replication genes, could affect both global synthesis and degradation. Perturbations on chromatin remodelers only specifically impaired the global synthesis rates but not the degradation rates, supporting the established theory that gene expression is regulated by chromatin folding. In addition to the enrichment of genes in transcription, spliceosome and mRNA surveillance, perturbation on OXPHOS genes and metabolism-related genes also affected the RNA processing, consistent with the fact that 5’ capping, 3’ polyadenylation, and RNA splicing are highly energy-dependent processes. That knockdown of OXPHOS genes and metabolism-related genes could reduce the mitochondrial transcriptome dynamics and also supported the complex feedback mechanisms between energy metabolism and mitochondrial transcription63. e-h. Scatterplots showing the relationships between dropout effects and global synthesis rates (e), global degradation rates (f), proportions of exonic reads in the nascent transcriptome (g), and mitochondrial RNA turnover (h). A linear regression line was fitted and ±95% confidence intervals are visualized for each metric. Dropout rank, the ascending rank of gene-level sgRNA counts log2FC from the bulk screen. Directions were assigned as shown in Figure 2e–h. Both global synthesis and degradation rates showed strong negative correlations with dropout, indicating knocking out essential genes generally resulted in impaired global RNA synthesis and degradation. In contrast, proportions of exonic reads in the nascent transcriptome were much more stable across perturbations, and were only specifically affected by genes functioning in RNA processing. Proportions of mitochondrial nascent reads were also prone to be affected by genetic perturbation, but directions of changes depend more on the functions of perturbed genes than the essentiality of genes.
