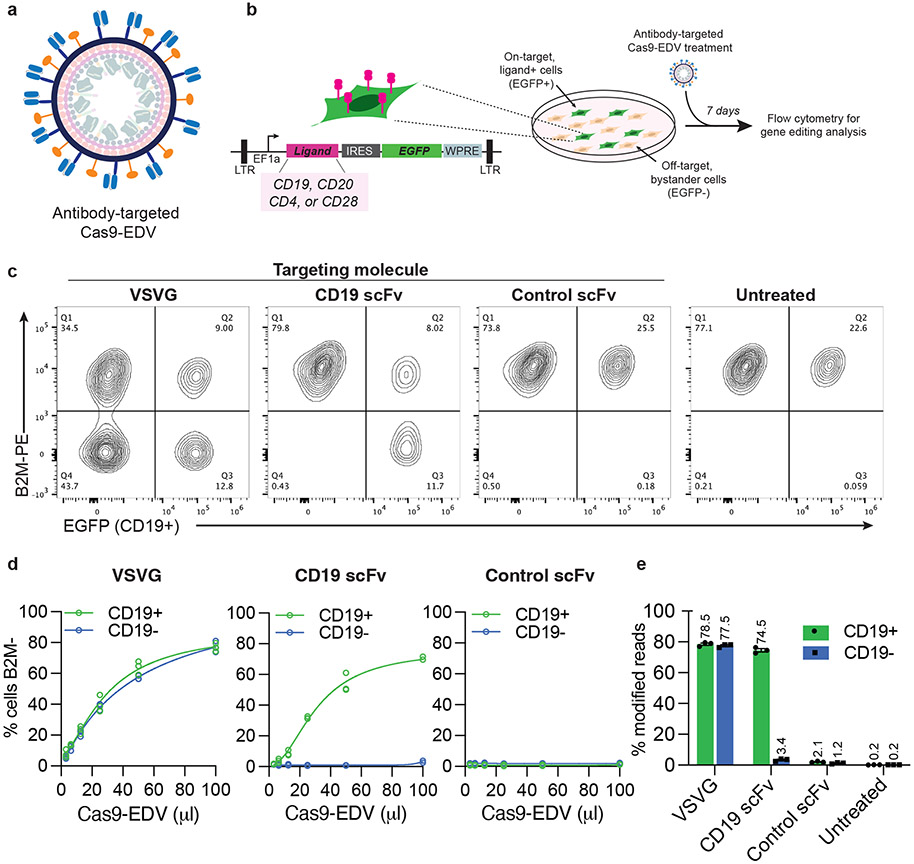
Fig. 1. Cell-specific genome editing with antibody-targeted Cas9-EDVs.
a, Schematic scFv targeting molecules (blue) and VSVGmut (orange) on the exterior surface of a Cas9-EDV. Cas9-EDVs package pre-formed Cas9-single guide RNA complexes to avoid genetically encoding genome editors within a viral genome. b, Experimental outline and schematic of the lentiviral vector used for engineering HEK293T EGFP cells that express heterologous ligands on the plasma membrane (e.g. CD19). To promote cellular engineering via single lentiviral integration events, engineered cell mixtures were generated via low multiplicity of infection to achieve <25% EGFP+ cells. Engineered cell mixtures were challenged with B2M-targeting Cas9-EDVs to test targeting molecule activity. c-e, Assessment of antibody-targeted Cas9-EDV activity. HEK293T and CD19 EGFP HEK293T cells were mixed at an approximate ratio of 3:1 and treated with B2M-targeting Cas9-EDVs displaying various targeting molecule pseudotypes. Cas9-EDVs were concentrated 10x, and cells were treated with 50 μl Cas9-EDVs (c, e) or in a dilution curve (d). Analysis was performed at 7 days post-treatment to assess B2M knockout in EGFP+ (on-target) and EGFP− (bystander) cells by flow cytometry (c, d) and amplicon sequencing (e). N=3 technical replicates were used in all experiments except for the 100 μl dose of CD19-scFv in (d) (N = 2). Individual replicate values and four-parameter non-linear regression curves are plotted (d). Error bars represent the standard error of the mean (e).