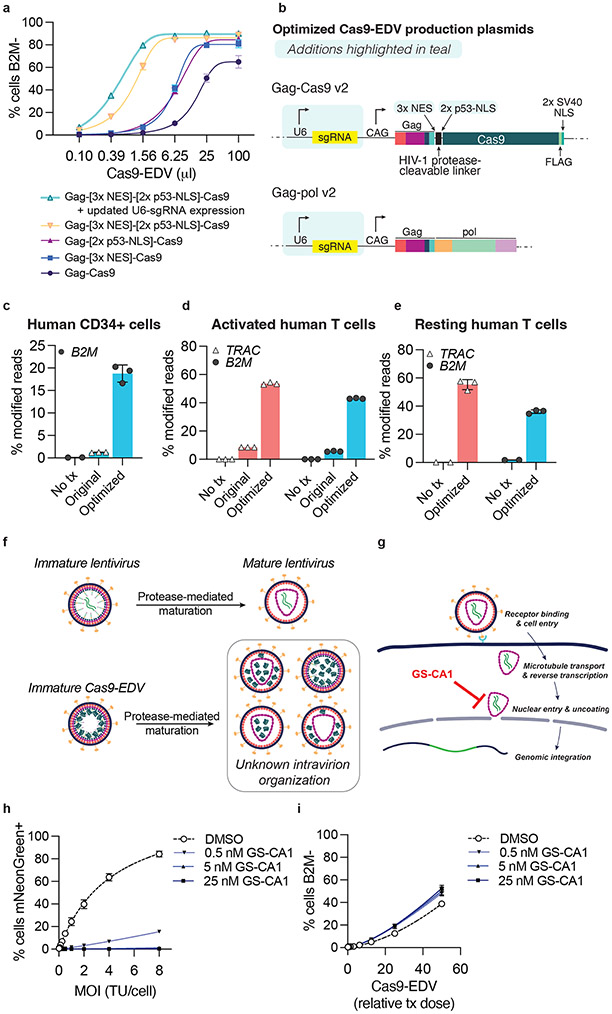
Fig. 2. Optimization of Cas9-EDVs for enhanced genome editing activity in primary human cells.
a, Genome editing activity comparison of CD19 antibody-targeted Cas9-EDV variants packaging B2M-targeted Cas9 ribonucleoproteins (RNPs). Expression of B2M protein was assessed by flow cytometry 7 days post-treatment in CD19-expressing target cells. b, Diagram of the optimized Gag-Cas9 and Gag-pol Cas9-EDV production plasmids; features updated from Hamilton & Tsuchida et al., 2021 are highlighted in teal. c-e, Genome editing activity of optimized VSVG-pseudotyped Cas9-EDVs in primary human CD34+ cells (c) and activated (d) and resting primary human T cells (e). B2M or TRAC genome editing was assessed by amplicon sequencing 7 days post-treatment. N=3 technical replicates were assessed for all conditions except for the untreated resting human T cells (N=2). f, Schematic of potential intra-particle Cas9-EDV configurations for packaged Cas9 RNPs following proteolytic maturation. g, Schematic of the compound GS-CA1 inhibiting either the nuclear import and/or uncoating of an HIV-1 capsid. h, An mNeonGreen lentiviral vector was used to transduce HEK293T cells at the indicated MOI in the presence of GS-CA1 or DMSO. The percent of mNeonGreen-positive cells was assessed by flow cytometry 3 days post-treatment. TU = transducing units. i, B2M-targeting Cas9-EDVs, pre-titered such that the highest treatment dose would result in approximately 50% cells B2M-, were used to transduce HEK293T cells in the presence of GS-CA1 or DMSO. B2M expression was assessed by flow cytometry 3 days post-treatment. Error bars represent the standard deviation of the mean. Unless otherwise noted, N=3 technical replicates were used in all experiments and four-parameter non-linear regression curves are plotted in a, h, i.