Fig. 3 |. Variation in the length and sequence composition of human centromeric α-satellite HOR arrays.
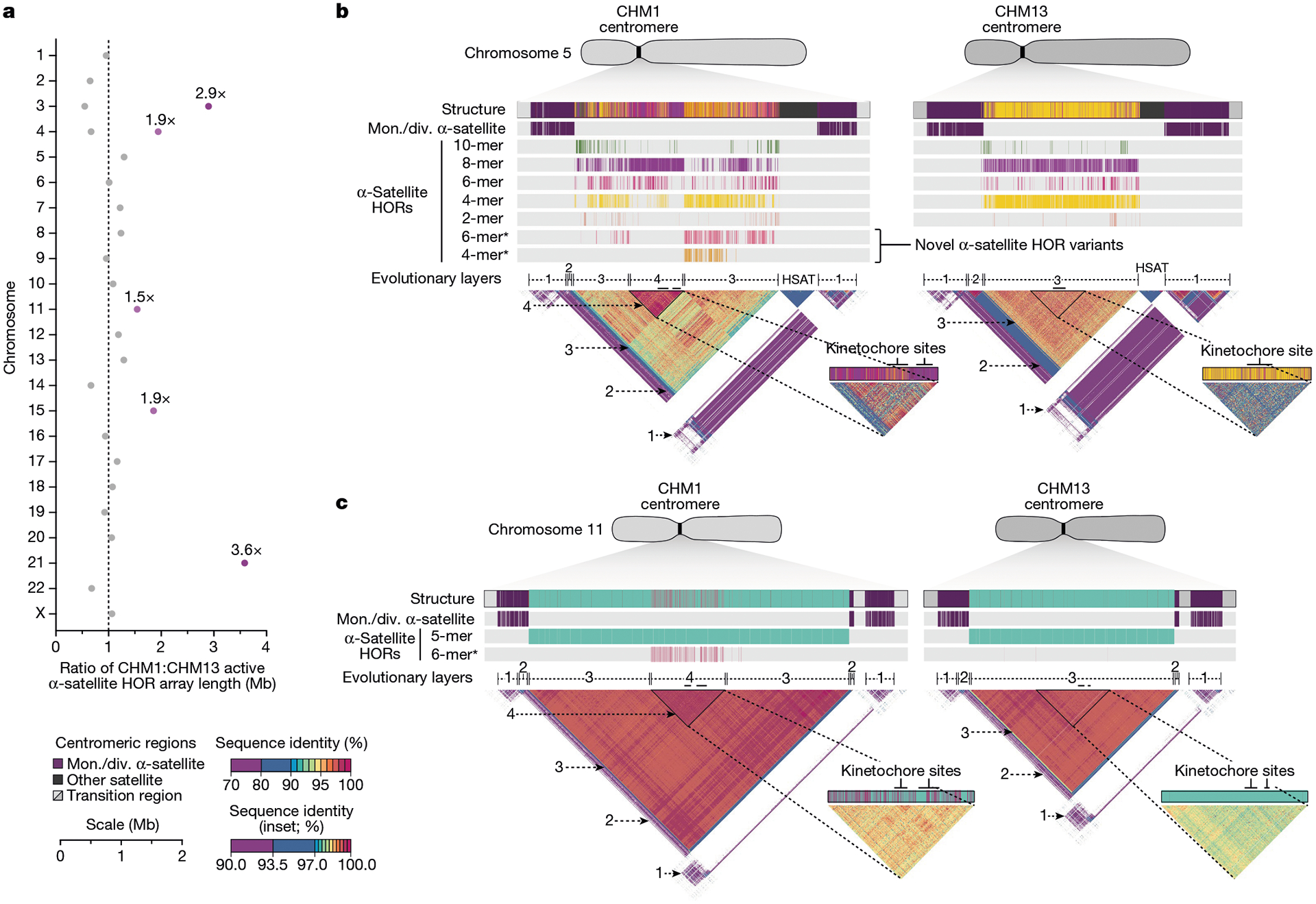
a, Ratio of the length of the active α-satellite HOR arrays in the CHM1 genome compared with those in the CHM13 genome. b,c, Comparison of the CHM1 and CHM13 chromosome 5 D5Z2 α-satellite HOR arrays (b) and CHM1 and CHM13 chromosome 11 D11Z1 α-satellite HOR arrays (c). The CHM1 chromosome 5 D5Z2 array contains two novel α-satellite HOR variants (Supplementary Fig. 44a) as well as a new evolutionary layer (layer 4; indicated by an arrow), which is absent from the CHM13 array. Similarly, the CHM1 chromosome 11 D11Z1 α-satellite HOR array contains a six-monomer HOR variant that is much more abundant than in the CHM13 array and comprises a new evolutionary layer, or a stretch of sequence that has evolved separately from neighbouring sequences (layer 4; indicated with an arrow), although this 1.21 Mb segment is more highly identical to the flanking sequence. The inset shows each of the new evolutionary layers with a higher stringency of sequence identity, as well as the relative position of the kinetochore. Notably, the α-satellite HOR variants comprising the new evolutionary layers in both CHM1 chromosomes 5 and 11 have divergent CpG methylation patterns despite their identical structure (Supplementary Fig. 74). Asterisk, α-satellite HORs variants that are either novel or present in higher abundance in the CHM1 centromere relative to the CHM13 centromere.
