Figure 2.
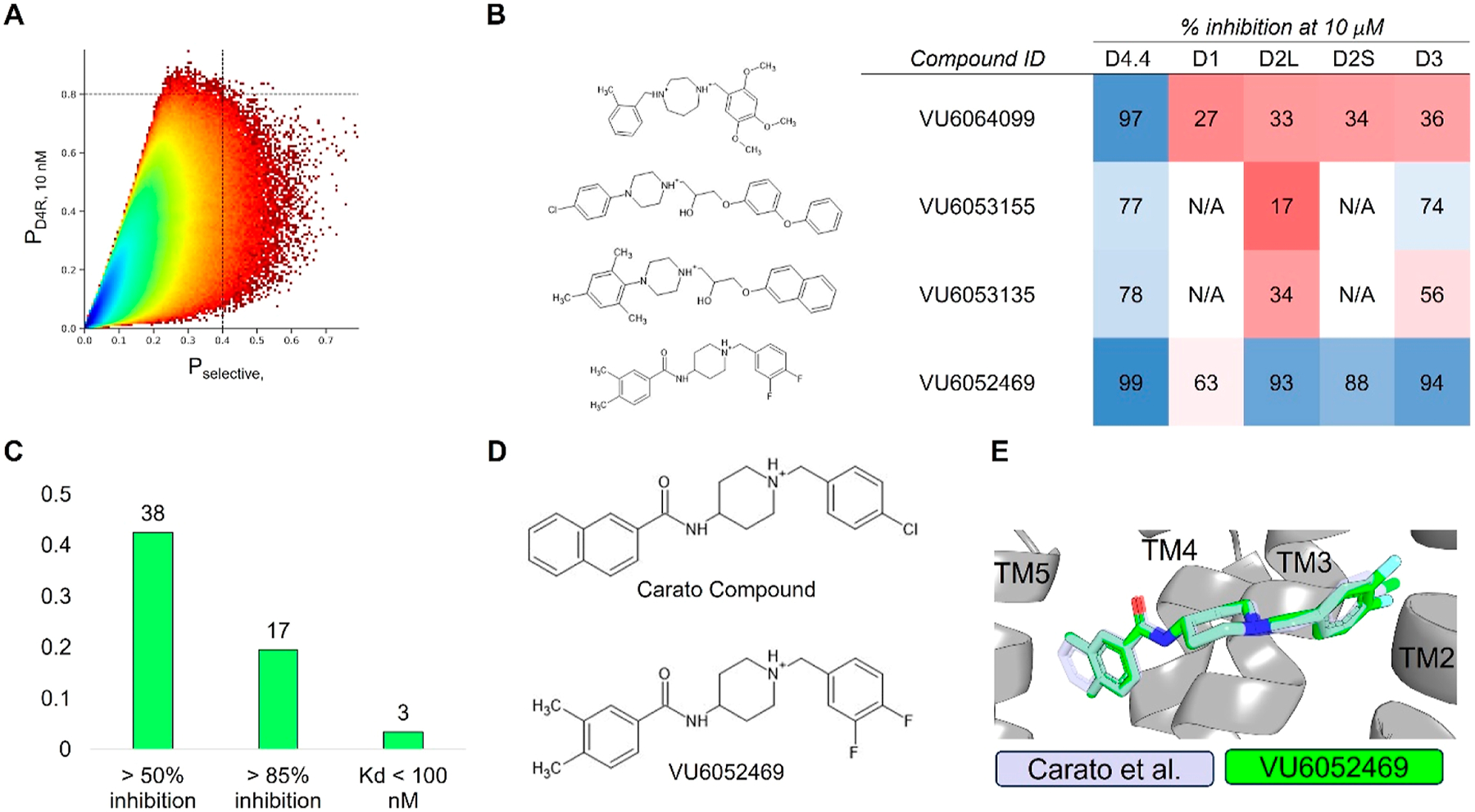
Virtual high-throughput screening for D4R antagonists. (A) Predicted D4R activity vs selectivity from the ligand-based multitask ANN QSAR model ultralarge library virtual high-throughput screening. Dashed lines indicate QSAR-predicted active classification probabilities at or greater than 80% (horizontal) and 40% (vertical) for D4R 10 nM activity and overall selectivity, respectively. Plot color is contoured by the density of molecules, with higher-density regions appearing blue and lower-density regions appearing red. (B) Sample molecules identified during the virtual high-throughput screening. (C) D4R hit-rate for experimentally validated molecules. (D) 2D structures of Carato et al.: compound 2271 and VU6052469. (E) Overlay of docked poses of Carato et al.: compound 2271 and VU6052469 within D4R.
