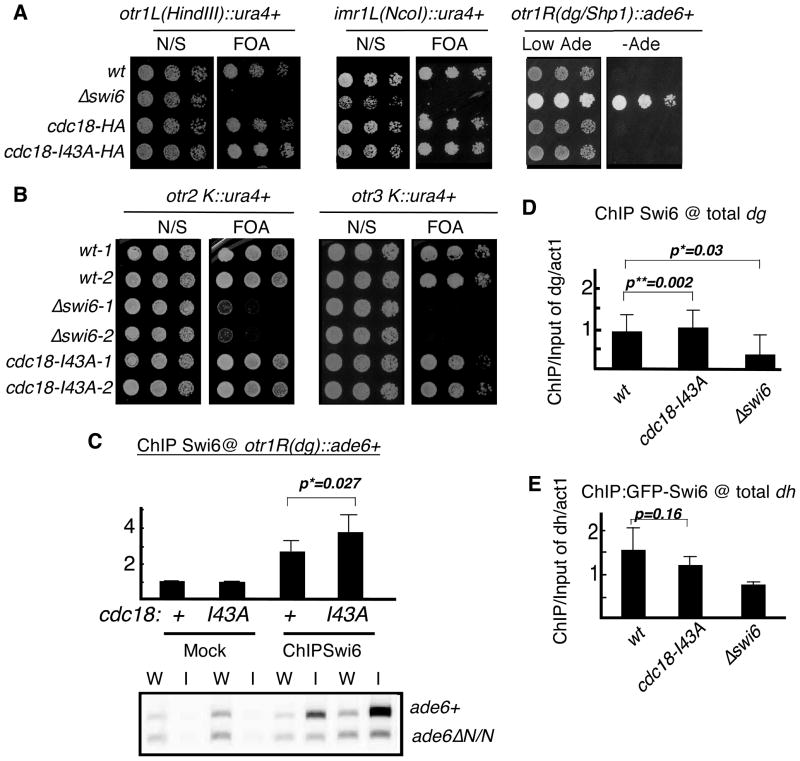
Fig 5. Region specific silencing defects in cdc18-I43A.
(A) Expression of the ura4+ or ade6+ marker at the cen1 dh, imr or dg repeat in cdc18-I43A mutant was determined by senstitivity to FOA or viability on minus adenine medium. N/S, non selective (Ura+) medium. (B) Expression of ura4+ marker in cen2 or cen3 dg repeat was determined by sensitivity to FOA. (C) Swi6 localization at transgenes in cen1 dg repeat (ade6+) in cdc18-I43A determined by ChIP. As an internal control, the same primers detect an ade6ΔN/N minigene at their normal euchromatic loci. Swi6 localization at global dg (D) or dh (E) repeat in cdc18-I43A mutant was determined by ChIP compared to act1. Swi6 in cdc18-I43A is significant higher at cen1(dg) and total dg regions, with p value of 0.027 and 0.002, but lower at dh regions. Statistical analysis was performed on 3 independent experimental samples using student t test.