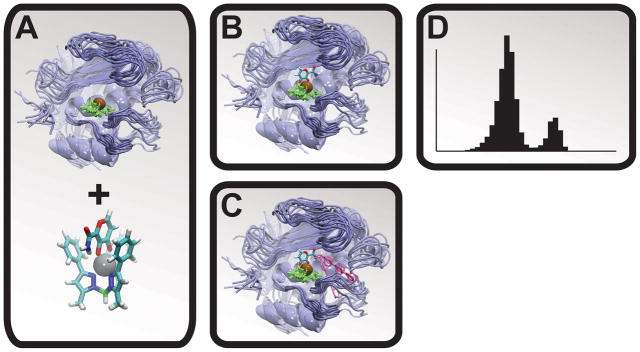
Figure 1.
The molecular-dynamics/docking protocol. The MMP protein is shown in blue, the active-site zinc cation is shown in brown, the zinc-coordinating histidine residues are shown in green, and representative LUDI-docked fragments are shown in pink. A) A molecular dynamics simulation was used to generate numerous MMP conformations. B) A model of the ZBG was positioned relative to the protein configurations by aligning a crystal structure of the bioinorganic model to the MMP active sites. C) LUDI was used to dock molecular fragments into the S1′ active sites. D) The resulting spectrums of LUDI scores were subsequently used to calculate ensemble-average scores.