Fig. 2.
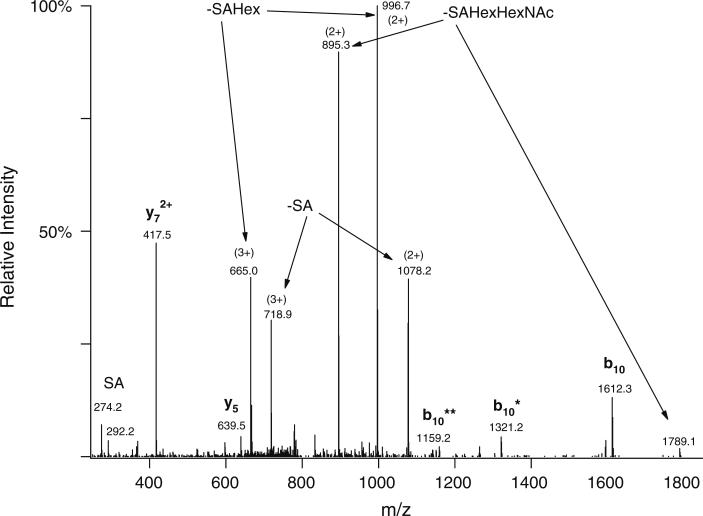
Linear ion trap CID spectrum of the same glycopeptide AVGAQVLESTPPPHVMR modified with SAGalGalNAc as in Fig. 1. Precursor ion was at m/z 815.7307(3+). The abundant peptide fragments in the spectrum were formed by cleavage N-terminal to the first Pro residue. The glycosylated half of the structure (b10) can be observed with the sugar attached as well as partially or completely deglycosylated—the number of asterisks indicates the number of carbohydrate units lost