Abstract
Current U.S. requirements for testing cell substrates used in production of human biological products for contamination with bovine and porcine viruses are U.S. Department of Agriculture (USDA) 9CFR tests for bovine serum or porcine trypsin. 9CFR requires testing of bovine serum for seven specific viruses in six families (immunofluorescence) and at least 2 additional families non-specifically (cytopathicity and hemadsorption). 9CFR testing of porcine trypsin is for porcine parvovirus. Recent contaminations suggest these tests may not be sufficient. Assay sensitivity was not the issue for these contaminations that were caused by viruses/virus families not represented in the 9CFR screen. A detailed literature search was undertaken to determine which viruses that infect cattle or swine or bovine or porcine cells in culture also have human host range [ability to infect humans or human cells in culture] and to predict their detection by the currently used 9CFR procedures. There are more viruses of potential risk to biological products manufactured using bovine or porcine raw materials than are likely to be detected by 9CFR testing procedures; even within families, not all members would necessarily be detected. Testing gaps and alternative methodologies should be evaluated to continue to ensure safe, high quality human biologicals.
Keywords: Adventitious agent testing, Animal cell culture, Viral contamination, Bovine virus, Porcine virus, Human host range
1. Introduction
Biological medicinal products for both human and veterinary use and the cell substrates used for their manufacture are currently tested for bovine and porcine adventitious viruses due to the widespread use of bovine serum and porcine or bovine trypsin in cell culture. Currently, testing for bovine and porcine adventitious agents is performed in compliance with the U.S. Department of Agriculture (USDA) regulations for veterinary products as specified in the 9CFR 113 regulations (9CFR). These regulations are intended to ensure that veterinary agents of concern are not introduced into new populations of animals (e.g., into new geographic regions or into previously unexposed herds or flocks), in order to prevent veterinary epidemics and protect the U.S. cattle and swine industries. Cell-culture derived vaccines for human use were developed in the 1950’s. Since fetal calf serum and bovine or porcine trypsin were used in cell culture, the 9CFR tests developed for veterinary use to screen for viruses that can infect cattle and swine were implemented by the authorities regulating human vaccines. However, many viruses not of significant concern to the cattle and swine industry are not addressed by the 9CFR testing. Today, over half a century after cell culture-derived vaccines were initially developed, the human biologics industry is still using the methods specified in the 9CFR regulations for testing FBS and porcine trypsin. When these tests are applied to bovine and porcine-derived raw materials used to manufacture products intended for human use, they may or may not detect the most relevant agents of concern for biological products for human use. The USDA regulations call for testing bovine serum for at least nine viruses; seven are tested specifically by immunofluorescence upon exposure and amplification in susceptible cells, and at least two more are tested by more general read-outs (hemagglutination/hemadsorption or cytopathic effect) upon exposure and amplification in susceptible cells. In addition, the USDA regulations require testing of porcine trypsin for porcine parvovirus by immunofluorescence upon exposure and amplification in susceptible cells.
In the context of the human biologicals industry, there are three main concerns with this approach. Firstly, it is not clear whether other known bovine or porcine viruses would likely be present in fetal bovine serum or porcine trypsin. Secondly, it has not been well considered which of these 9CFR-specified viruses have human host range. Thirdly, it has also not been systematically researched whether there are other bovine and porcine agents of equal or greater concern for human biologicals that are not specified or detected by the current testing.
To gather current relevant information to try and answer these questions, a research project was undertaken with the following goals:
To perform a literature search to define those viruses that could contaminate bovine and porcine raw materials used in production of human biologicals and which might pose a potential safety risk to recipients.
To evaluate current 9CFR testing for bovine and porcine viruses to predict whether those viruses defined as a potential risk to recipients of human biologicals can likely be detected by current 9CFR tests.
Serum and trypsin manufacturers use upstream sourcing controls and harvesting and pooling procedures to limit contamination of their products. However, these practices vary by manufacturer. In the absence of a consistent procedure for processing, the worst case in terms of potential contamination was assumed; i.e., that any virus worldwide that could contaminate bovine serum or porcine trypsin and has human host range is a potential contaminant. While this assumption is a stringent one, in fact, risk mitigation procedures can and should begin at the manufacturer of the animal-derived material and at the collection abattoirs. Regulatory agencies expect manufacturers of human biologicals to regularly audit their suppliers of serum, trypsin, and other animal-derived materials, to assure that proper risk mitigation procedures are followed.
Serum collection is a non-sterile procedure and the blood from 2500 to 3000 fetuses is used to produce a single 1500 liter lot of serum [1]. Similarly, the production of porcine trypsin uses huge numbers of pancreases that are extracted to produce a lot (batch) of trypsin. One infected animal may contaminate an entire batch/lot and the sensitivity of current testing may not be adequate to detect diluted contaminants. The limitations of testing for viruses in pooled products have been amply demonstrated in the human blood industry, and introduction of the mini-pool testing approach has led to increased sensitivity of detection and thereby increased safety for human recipients of these products. An additional safety concern for bovine serum and porcine trypsin is whether equipment cleaning between processing of animal-derived material lots is adequate to prevent cross-contamination.
It should be noted that the lots of bovine serum and trypsin used to produce human biologicals are often subjected to virus removal and inactivation procedures, such as filtration, heat inactivation or gamma-irradiation, before use in manufacturing biological products for human use. What is unclear is whether the testing for adventitious viruses performed by the vendor or by the manufacturer using the materials is always conducted before the use of inactivation procedures, when the likelihood of detecting a contaminant would be higher, or whether it may occur after treatment, when detection would be significantly less likely due to the inactivation of the majority, but possibly not all, of the contaminant.
Many newer products are cell-based or utilize viral vectors and are more like traditional live-attenuated vaccines where manufacture provides less opportunity to remove/inactivate viral contaminants.
There have been several well-publicized contamination events caused by bovine/porcine agents that provide the opportunity to evaluate our current virus safety approach:
Cache Valley Virus (CVV) –fermentors from multiple manufacturers have been infected. CVV, although not typically recognized as a bovine virus, is a multispecies virus with bovine host range [2].
Calicivirus 2117 [vesivirus]-facility contamination forced a costly facility shutdown resulting in product shortage that deprived patients of product [3].
Multiple contaminations of fermentors with reoviruses, another virus family with wide host range [4].
Porcine circovirus type 1 and type 2 nucleic acid contamination of live rotavirus vaccines [5, 6].
Contaminations of cell cultures during production of biological products often arise when huge scale-up procedures using large quantities of bovine serum or porcine trypsin are needed, and where the sensitivity of the tests currently being used is not adequate to detect a contaminant that starts at a low-level and amplifies over the long course of cell culture. Contamination has also been detected when new testing methods are applied, such as for porcine circovirus (PCV).
It is important to recognize that direct testing of bovine serum and porcine trypsin is not the only measure in place for assuring product safety and is used in conjunction with testing for viruses in cell banks and lot-to-lot in-process testing of unprocessed bulk harvest material. However, these other tests are not designed to specifically detect bovine or porcine viruses. Additionally, for some products, removal and inactivation of viruses during purification procedures must be demonstrated. This report will focus on bovine and porcine agents and their ability to be detected by the 9CFR procedures currently in use.
2. Methods
At the start of the literature search, the following goals were developed to guide and focus the search: (i) to determine whether the seven bovine viruses specified in 9CFR 113.47 [BVDV, REO, Rabies, BTV, BAd, bovine parvovirus and BRSV] are capable of infecting humans or display human host range (e.g., by infecting human cells in culture or producing antibodies in humans), (ii) to determine if, in addition to the 7 specific bovine viruses, there are other bovine viruses that would be detected by the hemadsorption/hemagglutination and cytopathic effect (CPE) procedures in 9CFR, (iii) to identify porcine viruses (in addition to porcine parvovirus) that have human host range and that could contaminate porcine trypsin, and (iv) if additional porcine viruses were identified, to determine whether the 9CFR procedure for testing of porcine cells [9CFR113.47] would detect them.
At the outset, the pathogen search sought to identify all known vertebrate viruses with bovine or porcine host ranges [natural infection, or detection of antibodies, or ability to grow in bovine or porcine cells in culture]. The second criterion for inclusion was that each virus identified also had to have human host range, which includes more than traditionally zoonotic viruses. Searching for zoonotic bovine and porcine viruses was assumed not to be adequate for this study because biological products are frequently injected, bypassing normal host immune responses. Therefore, human host range was defined by the presence of any of the following: natural infection [including laboratory accidents], detection of antibodies (with or without disease manifestation), or the ability to infect human cells in culture. These were defined as Agents of Concern.
Literature on viruses with human host range was also analyzed for infectivity among species. Bovine viruses were analyzed for the potential to infect pigs, and porcine viruses were analyzed for the potential to infect cattle.
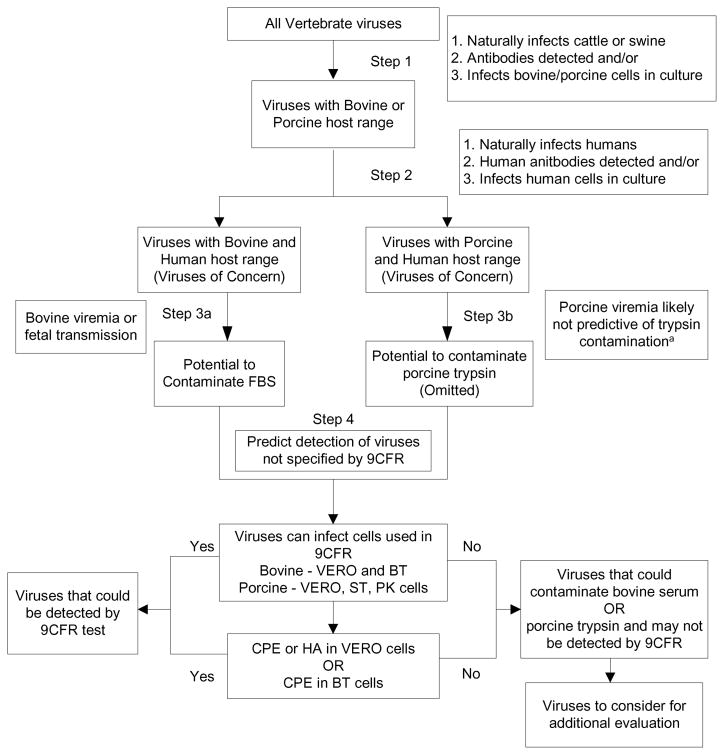
A conservative approach was adopted to ensure viruses posing a potential threat were not inadvertently eliminated from consideration due to uninvestigated properties. The step-wise analysis used is detailed in Figure 1 below.
Figure 1. Schematic of Step-Wise Analysis.
This process was used to identify viruses with bovine and porcine host range also exhibiting human host range, that could contaminate source materials, and that may not be detected by 9CFR testing
a Data found shows that porcine viremia is likely not predictive of pancreatitis (section 3.2)
3. Results
3.1 Viruses with Human Host Range
The first analysis step identified viruses with bovine or porcine host range and step 2 selected those viruses that also display human host range. These included documented zoonotic viruses, laboratory infections after accidental exposure, detection of antiviral antibodies, and virus infection of human cells in vitro. The virus list was then narrowed to include only viruses with bovine or porcine host range that also display human host range [Tables 1 and 2]. One difficulty was that often information was obtained in reference to a genus or group of viruses (e.g., hantaviruses) but specific information on individual viruses was lacking. Also, some viruses, such as Chikungunya and Powassan, were present in vectors (mosquitoes and ticks) that feed on cattle but no direct evidence of bovine host range could be found. A virus such as porcine lymphotropic herpesvirus (PLHV), although a concern for xenotransplantation of porcine tissues into humans, was not included in the tables since it does not meet the criteria for human host range.
Table 1.
Viruses of Concern with Bovine and Human Host Range
| Virus of Concern |
|---|
| Adenoviridae |
| Bovine adenovirus BAdV-9 = Human adenovirus C [7] |
| Anelloviridae (proposed family) |
| Torque teno virus TTV [8] |
| Bornaviridae |
| Borna disease virus BDV [9–12] |
| Bunyaviridae |
| Aino virus [13, 14] |
| Cache valley virus CVV [2] |
| Crimean Congo haemorrhagic fever virus CCHF [15] |
| Hantaan virus HTNV [16–18] |
| Jamestown Canyon virus JCV [19, 20] |
| LaCrosse virus LACV [19, 21] |
| Puumala virus [18, 22] |
| Rift valley fever virus RVFV [23] |
| Caliciviridae |
| Norovirus [formerly Norwalk agent] [24, 25] |
| San Miguel sea lion virus SMSV-5 [26] |
| Circoviridae |
| Bovine circovirus BCV = evolved strain of Porcine circovirus type 2 PCV-2 [27, 28] |
| Coronaviridae |
| Bovine coronavirus BCoV-1 [29] |
| Bovine torovirus BtoV [30, 31] |
| Flaviviridae |
| Bovine viral diarrhea virus BVDV [32, 33] |
| Japanese encephalitis virus JEV[34] |
| Kyasanur forest disease virus KFDV [34] |
| Louping ill virus [34, 35] |
| Murray Valley encephalitis virus MVE [34] |
| Saint Louis encephalitis virus SLEV [34, 36] |
| Tick borne encephalitis virus TBEV[37 ] |
| Wesselsbron virus [34] |
| West Nile virus ( including Kunjin) [38] |
| Hepeviridae |
| Hepatitis E virus HEV [39–41] |
| Herpesviridae |
| Bovine herpesvirus BHV-4 [MOVAR] [42, 43] |
| Equine herpesvirus EHV-1 [44, 45] |
| Infectious bovine rhinotracheitis virus IBR= BHV-1 [46] |
| Pseudorabies virus PRV [47, 48] |
| Orthomyxoviridae |
| Dhori virus [49] |
| Influenza A virus [50], Avian influenza virus [51] |
| Thogotovirus THOV [49, 52] |
| Papillomaviridae |
| Bovine papilloma virus BPV [53] |
| Paramyxoviridae |
| Bovine parainfluenza virus BPIV3 [54, 55] |
| Bovine respiratory syncytial virus BRSV [56] |
| Peste-des-petits ruminants virus PPRV [57, 58] |
| Rinderpest virus RPV [59, 60] (Declared eradicated by the United Nations in 2010) |
| Parvoviridae |
| Bovine adeno-associated virus BAAV [61] |
| Bovine hokovirus BHoV [62] |
| Picornaviridae |
| Bovine enterovirus BEV-1, BEV-2 [63] |
| Bovine kobuvirus BKV-1 U-1 strain [64, 65] |
| Encephalomyocarditis virus EMC [66, 67] |
| Foot and mouth disease virus FMDV [68, 69] |
| Seneca valley virus SVV [70] |
| Polyomaviridae |
| Bovine polyomavirus BPyV [71] |
| Poxviridae |
| Aracatuba virus [72] |
| Bovine papular stomatitis virus BPSV [73] |
| Cantagalo virus [74] |
| Cowpox virus [15] |
| Pseudocowpox virus PCPV [75] |
| Vaccinia virus [76] |
| Reoviridae |
| Banna virus BAV [77, 78] |
| Bluetongue virus BTV [79] |
| Epizootic haemorrhagic disease virus EHDV [80, 81] |
| Liao Ning virus LNV [82] |
| Reovirus [83, 84] |
| Rotavirus [65, 85, 86] |
| Retroviridae |
| Bovine foamy virus BFV [87] |
| Bovine leukemia virus BLV [88–90] |
| Rhabdoviridae |
| Bovine ephemeral fever virus BEFV [91] |
| Rabies virus [92] |
| Vesicular stomatitis virus VSV [15, 93, 94] |
| Togaviridae |
| Eastern equine encephalitis virus EEEV [95, 96] |
| Getah virus [97, 98] |
| Ross River virus RRV [97, 99] |
| Sindbis virus [97] |
| Venezuelan equine encephalomyelitis virus VEE [100, 101] |
Table 2.
Viruses of Concern with Porcine and Human Host Range
| Virus of Concern |
|---|
| Anelloviridae (proposed family) |
| Torque teno virus TTV [8] |
| Bunyaviridae |
| Crimean Congo haemorrhagic fever virus |
| CCHF [15] |
| Hantaan virus HTNV [16, 17] |
| Jamestown Canyon virus JCV [19, 21] |
| LaCrosse virus LCV [19, 21] |
| Caliciviridae |
| Norovirus [formerly Norwalk agent] [24, 25] |
| San Miguel sea lion virus SMSV-5 [26] |
| Sapovirus [102] |
| Circoviridae |
| Porcine circovirus PCV-1 & PCV-2 [27, 103] |
| Coronaviridae |
| Bovine coronavirus BCoV-1 [104, 105] |
| Severe acute respiratory syndrome virus SARS [106, 107] |
| Transmissible gastroenteritis virus TGEV [108] |
| Filoviridae |
| Ebola Reston virus [109] |
| Flaviviridae |
| Bovine viral diarrhea virus BVDV [32] |
| Dengue virus [110] |
| Ilheus virus [34] |
| Japanese encephalitis virus JEV [34] |
| Louping ill virus [34, 35] |
| Murray Valley encephalitis virus MVE [111] |
| Powassan virus [48, 112] |
| Tick borne encephalitis virus TBEV [113] |
| Wesselsbron virus [34] |
| West Nile virus WNV (including Kunjin) [114] |
| Hepeviridae |
| Hepatitis E virus HEV [39, 40] |
| Herpesviridae |
| Infectious bovine rhinotracheitis virus IBR= BHV-1 [115] |
| Porcine cytomegalovirus PCMV (B. Potts personal communication) |
| Pseudorabies virus PRV [47, 48] |
| Orthomyxoviridae |
| Avian influenza virus (H5N1), Porcine influenza virus (H1N1, H1N2) [116] |
| Paramyxoviridae |
| Bovine parainfluenza virus BPIV3 [54, 55, 117] |
| Menangle virus MENV [118–120] |
| Nipah virus NiV [121–124] |
| Peste-des-petits ruminants virus PPRV [57, 125] |
| Rinderpest virus RPV [126] (Declared eradicated by the United Nations in 2010) |
| Tioman virus TIOV [127] |
| Parvoviridae |
| Porcine hokovirus PHoV [62] |
| Porcine parvovirus PPV [128] |
| Picornaviridae |
| Encephalomyocarditis virus EMC [66] |
| Foot and mouth disease virus FMDV [68, 69] |
| Porcine enterovirus PEV-9 PEV-10 [129] |
| Seneca valley virus SVV [70] |
| Swine vesicular disease virus SVDV [130] |
| Reoviridae |
| Banna virus BAV [77, 78] |
| Reovirus [83, 84] |
| Rotavirus [65, 85, 86] |
| Retroviridae |
| Porcine endogenous retrovirus PERV [131] |
| Rhabdoviridae |
| Rabies virus [92] |
| Vesicular stomatitis virus VSV [15, 93, 94] |
| Togaviridae |
| Eastern equine encephalitis virus EEEV [132] |
| Getah virus [98] |
| Ross River virus RRV [99] |
| Venezuelan equine encephalomyelitis VEE [133] |
Note: Closely related viruses having human host range that are in the same family as those listed in either table above were included in the database and may also be of concern but were omitted from the table above because specific information relating to bovine or porcine host range was not found.
Expanding the definition of human host range beyond documented zoonosis resulted in approximately a 10-fold increase in the number of viruses with bovine and porcine host range (Tables 1 and 2) that are not included in the agents currently screened by 9CFR. Many of these viruses are appropriately not of concern to the cattle and swine industry.
3. 2 Potential Source Material Contaminants
Next, viruses on the list were evaluated for their potential to contaminate source materials used to produce biological products. In the absence of direct reports of detection of a particular virus in bovine serum, reported viremia or fetal transmission were used as surrogates for predicting the potential to contaminate serum. These markers identify the potential to cross the placenta, to accidentally contaminate FBS. Fetal transmission was defined as actual or experimental transmission of the agent or of a closely related agent.
Viremia could not be used as a surrogate for prediction of the potential to contaminate porcine trypsin since two porcine viruses reported to cause pancreatitis, transmissible gastroenteritis virus (TGEV) and porcine hemagglutinating encephalomyelitis coronavirus (PHE-CoV), were also reported not to cause viremia. Therefore, most porcine viruses with human host range were included as potential contaminants of porcine trypsin or porcine cells.
3.3 Predicted Detection of Viruses by 9CFR
The list of the agents of concern was then evaluated to predict whether these viruses might be detected by 9CFR testing. For those viruses not detectable by 9CFR testing, alternative methods may need to be identified or developed. The first criterion for possible detection by 9CFR is the capability to grow in tissue culture.
Detection of Bovine Viruses by the 9CFR Procedure
The 9CFR methods to detect bovine agents (9CFR113.47/.53) utilize monolayers of African green-monkey kidney (VERO) cells or bovine turbinate (BT) cells. The tests are run for 21 days, and the cells are observed for (CPE) and hemadsorption [HAd] or hemagglutination of erythrocytes [HA]. At 21 days, the cells are subjected to immunofluorescence assay (IFA) using antisera specific for seven viruses:
bluetongue virus
bovine adenoviruses
bovine parvovirus
bovine respiratory syncytial virus
bovine viral diarrhea virus (BVDV)
rabies virus
reovirus (REO-3 is generally used as the positive control in the assay, but the sensitivity of the assay for detection of REO-1 and REO-2 is not clear)
The virus list in Table 1 was analyzed for viruses reported to cause CPE, HAd, or HA in VERO or BT cells. This approach permitted analysis of those bovine viruses that are not deliberately included in the 9CFR testing but have a high likelihood to be detected by 9CFR testing. The bovine viruses with human host range that are likely not to be detected by 9CFR testing are reported in Table 3. For a more conservative estimate, viruses that had documented infection of and/or isolation in the cell lines used in 9CFR testing were included. Viruses that produced CPE on VERO E6 cells, a subclone of VERO used by some testing laboratories, were also considered likely to be detected, but this would need to be confirmed through direct testing.
Table 3.
Viruses with Human and Bovine Host Range that are Not Predicted to be Detected by 9CFR Test
| Viruses to Consider for Additional Evaluation |
|---|
| Anelloviridae (proposed family) |
| Torque teno virus TTV* |
| Bornaviridae |
| Borna disease virus BDV [134] |
| Bunyaviridae |
| Cache valley virus CVV [2] |
| Puumala virus [135] |
| Caliciviridae |
| Norovirus [formerly Norwalk agent] [136] |
| Circoviridae |
| Bovine circovirus bovCV* |
| Coronaviridae |
| Bovine torovirus BtoV* |
| Flaviviridae |
| Louping ill virus* |
| Saint Louis encephalitis virus SLEV* |
| Wesselsbron virus [137] |
| Hepeviridae |
| Hepatitis E virus HEV* |
| Papillomaviridae |
| Bovine papilloma virus BPV* |
| Parvoviridae |
| Bovine adeno-associated virus BAAV* |
| Bovine hokovirus BHoV* |
| Picornaviridae |
| Bovine enterovirus BEV-1*, BEV-2* |
| Seneca valley virus SVV [138] |
| Polyomaviridae |
| Bovine polyomavirus BPyV [71, 139] |
| Poxviridae |
| Aracatuba virus* |
| Cantagalo virus* |
| Cowpox virus [140] |
| Pseudocowpox virus PCPV [141, 142] |
| Reoviridae |
| Banna virus BAV [82] |
| Epizootic haemorrhagic disease virus EHDV [143] |
| Rotavirus [144] |
| Retroviridae |
| Bovine foamy virus BFV [145] |
| Bovine leukemia virus BLV* |
| Togaviridae |
| Ross River virus RRV [99] |
Isolation versus growth in the cells used in 9CFR is critical, since some adapted viruses grow readily in VERO, but original isolates are difficult to grow or cannot be isolated in VERO. Although some predictions are possible, appropriate validation that the 9CFR test can actually detect field isolates of these viruses would be critical to reliance on the 9CFR test as a valid safety test for contamination of raw materials with these agents.
Detection of Porcine Viruses by the 9CFR Procedure
The 9CFR methods to detect porcine agents [9CFR113.53(d)] in porcine trypsin use VERO, porcine kidney (PK), and swine testicular (ST) cells. Because trypsin is known to inactivate many viruses, porcine trypsin is only tested for porcine parvovirus. The testing utilizes inoculation of cells of proven equal sensitivity to PPV for 14 days with at least 1 subculture followed by IFA for PPV.
When following the 9CFR methods for porcine cells, for example, in xenotransplantation, the cell cultures are observed for CPE and tested for hemadsorption. At the end of culture, the cells are subjected to IFA using antisera specific for six viruses:
bovine viral diarrhea virus (BVDV)
rabies virus
reovirus
porcine adenovirus
transmissible gastroenteritis virus (TGEV)
porcine hemagglutinating encephalomyelitis virus (PHE-CoV)
Since a 9CFR procedure exists for detection of porcine viruses for use with porcine products other than trypsin, an assessment was performed for porcine viruses in Table 2 and their ability to be detected in VERO, VERO E6, ST or PK cells used in the 9CFR testing of porcine cells. Viruses that are inactivated by trypsin were eliminated from the list of porcine viruses under the assumption they would not pose a risk to human health after inactivation. The porcine viruses with human host range that are likely not to be detected by 9CFR testing are reported in Table 4.
Table 4.
Viruses with Human and Porcine Host Range that are Not Predicted to be Detected by 9CFR Test
| Viruses to Consider for Additional Evaluation |
|---|
| Anelloviridae (proposed family) |
| Torque teno virus TTV* |
| Caliciviridae |
| Norovirus [formerly Norwalk agent] [136] |
| Sapovirus* |
| Circoviridae |
| Porcine circovirus PCV-1 & PCV-2 [146](B.Potts personal communication) |
| Flaviviridae |
| Louping ill virus[133] |
| Powassan virus [147] |
| Wesselsbron virus [137] |
| Hepeviridae |
| Hepatitis E virus HEV* |
| Herpesviridae |
| Porcine cytomegalovirus PCMV * |
| Paramyxoviridae |
| Menangle virus MENV [119] |
| Parvoviridae |
| Porcine hokovirus PHoV [62] |
| Picornaviridae |
| Porcine enterovirus PEV-9 PEV-10 [129] |
| Seneca valley virus SVV [138] |
| Reoviridae |
| Banna virus BAV [82] |
| Rotavirus [144] |
| Retroviridae |
| Porcine endogenous retrovirus PERV* |
| Togaviridae |
| Ross River virus RRV [99] |
Note: These are not exhaustive lists, and other viruses related to those listed above may also be of concern.
No information found to allow assessment of ability to be detected by 9CFR, hence considered as potential risk
Searches performed to determine whether individual porcine viruses can infect human cells revealed that 33 of the 55 porcine viruses with human host range can replicate in VERO cells. This information means that many, but not all of the porcine viruses of concern likely can be cultured in either a porcine cell line or in VERO cells. As indicated above, verification that the tests actually detect field strains in a validated assay is important. Additional viruses detectable using a 21 day 9CFR 113.47 test for porcine cells include bluetongue virus (BT), rotavirus, porcine reproductive and respiratory syndrome virus (PRRS), pseudorabies virus (PRV), JEV, swine encephalitis virus, and swine influenza [148]. The results of this analysis are incorporated in Table 5.
Table 5.
Table of virus families for which one or more member of the virus family given in the row have the characteristic in the given column heading, indicated by shaded boxes.
| Virus family | Bovine | Porcine | IFA read out in 9CFR testing | ||||
|---|---|---|---|---|---|---|---|
| Viruses with HHR | Capable of fetal transmission | May contaminate FBS | Predicted to cause CPE or HAd in 9CFR | Viruses with HHR | Predicted to cause CPE or HAd in 9CFR | ||
| Adenoviridae | |||||||
| Anelloviridae [proposed family name] | |||||||
| Bornaviridae | |||||||
| Bunyaviridae | |||||||
| Caliciviridae | |||||||
| Circoviridae | |||||||
| Coronaviridae | |||||||
| Filoviridae | |||||||
| Flaviviridae | |||||||
| Hepeviridae [proposed family name] | Unknown | ||||||
| Herpesviridae | |||||||
| Orthomyxoviridae | |||||||
| Papillomaviridae | Likely | ||||||
| Paramyxoviridae | |||||||
| Parvoviridae | |||||||
| Picornaviridae | |||||||
| Polyomaviridae | |||||||
| Poxviridae | |||||||
| Reoviridae | |||||||
| Retroviridae | |||||||
| Rhabdoviridae | |||||||
| Togaviridae | Unknown | ||||||
Note: This is not a comprehensive list of vertebrate virus families, only bovine or porcine viruses that were considered to have human host range were analyzed.
Since porcine cells are proposed for use in xenotransplantation, information found on porcine viruses other than PPV is also presented here. As mentioned above in 3.1, a virus such as PLHV is a concern for xenotransplantation of porcine derived tissues but does not have demonstrated human host range so is not listed in the tables but should be considered for further investigation. Additional products for which porcine viruses are a concern include human enzyme replacement pancreatic products. Given the recent finding of porcine circovirus nucleic acid in live rotavirus vaccines [6, 27] transmission of porcine viruses other than parvovirus from trypsin may be a broader concern than previously considered.
As in the case with bovine viruses [3.2.2], the literature contains many studies on HA using goose erythrocytes rather than chicken, and often the temperatures studied differed from those specified in 9CFR.
3.4 Analysis by Virus Family
Since many viruses that were identified were closely related and virus properties are important for virus clearance using solvent/detergent or filtration, viruses were also grouped into virus families. For bovine viruses with human host range, 21 virus families are represented while for porcine viruses, 17 virus families are represented [Table 5]. Shaded boxes in the table indicate that one or more members of the virus family in the given row have the characteristic indicated in the given column heading.
3.5 Assessment of viruses that may be inactivated by trypsin
Many viruses are inactivated by the enzymatic activity of trypsin. PPV is resistant to trypsin. Other viruses, like rotaviruses, are activated by low concentrations of trypsin. The starting material for trypsin preparation [i.e., the pancreatic extract] likely contains other porcine pancreatic enzymes [nucleases, glycosylases, lipases, etc.] that could contribute to virus inactivation. The extent of inactivation would likely vary depending on the method used for preparation of the trypsin, and therefore the virus contamination would likely vary from one trypsin preparation and one manufacturer to another. Trypsin preparations commonly used in biologics manufacturing are prepared by fractionation and chromatographic methods that result in relatively high purity but the presence of other enzymes is possible.
4. Discussion
One of the original goals of this study was to assess whether the current 9CFR testing is sufficient to ensure the safety of human biologicals. It is clear from the analyses performed that there are gaps between those viruses predicted to be detected by the 9CFR tests and those viruses that may have human host range and thus, may be of concern as contaminants. Also, in assessing the ability to be detected by 9CFR, it became apparent that many virus strains currently in virus culture collections were originally isolated by adaptation to cell culture. The ability of virus field isolates to grow in VERO cells within the time and passage constraints of the 9CFR assay needs to be explored.
There are virus families for which one or more members would likely be detected, but other members may not. In addition, the IFA read-outs are specific for members of a virus type (e.g., within bovine adenoviruses, the IFA is performed for only one of several types). Based on our literature search, our recommendation would be to retain the current 9CFR testing regimen for testing of FBS and supplement with additional testing procedures [e.g., PCR, see below]. In terms of modifying 9CFR testing of human biologicals, one may consider removing the IFA testing of bovine parvovirus and bovine RSV. BPV is a bocavirus that is not known to exhibit human host range. However, there are two other bovine parvoviruses [BAAV and bovine hokovirus] that are of concern and unlikely to be detected in the 9CFR test, so a general test for parvoviruses may be useful. BRSV was once thought to be a single virus that infected cattle and humans but now BRSV and HRSV are known to be separate viruses. BRSV can infect human cell lines in culture so it could be a risk for products produced in human cell lines (e.g., MRC-5, HeLa, HEK 293, PER.C6), although it is not known to cause human disease.
For testing of porcine trypsin, testing using 9CFR for porcine parvovirus should continue; however, a 21d test should be considered. For testing of other products derived from porcine cells, such as some gene therapy products, the specific testing for PHE-CoV could be removed.
Below, we provide a number of options to be implemented separately or together, regarding how to address the issue of viruses known or predicted not to be detected by current 9CFR tests but of potential concern to human health were they to contaminate human biologicals made using bovine serum or porcine trypsin.
Options:
4.1. No change in current practices and testing
The current testing algorithm is an orthogonal approach consisting of testing raw materials directly, testing bulk products for adventitious agents, monitoring of manufacturing processes for unexpected results (e.g. fermentor failures) and evaluation of the clearance of specific viruses by the manufacturing processes. This approach seems to have, for decades, prevented contamination that could have resulted in human disease. The recent finding of PCV genomic DNA in live rotavirus vaccines is of concern for product purity and quality. Although there is no evidence that the viral DNA represents a threat to human health [6, 27], PCV1 and PCV2 have been shown to infect human cells in culture [27]. In addition to the current testing algorithm, current Good Manufacturing Practices aid in ensuring product purity and quality, as well as manufacturing consistency, and as a result of these practices all of the contaminations discussed earlier, with the exception of PCV, were caught before product was released to the market.
4.2. Modify 9CFR Testing
Regulators of human biologicals should consider whether it would be appropriate to omit tests for viruses not of concern to human health and supplement with testing for specific additional viruses, such as bunyaviruses and reoviruses in addition to reovirus type 3, which are a concern to human health.
4.3. Require testing of bovine sera for bovine antibodies
In addition to possible cross-contamination with bovine serum during collection, the high price of fetal calf serum has the potential to encourage deliberate dilution of FBS with bovine or newborn calf serum. Another concern is the possible deliberate spiking with serum obtained from unapproved countries of origin. Maternal antibodies do not cross the bovine placenta, so a bovine fetus should not have maternal antibodies, and instituting testing for antibodies that would not be expected to be present in the fetus could address some of this concern.
4.4. Require gamma irradiation of serum and/or use of recombinant trypsin or serum-free media formulations
For serum
Manufacturers should continue to be encouraged to use gamma irradiation or other highly effective inactivation procedures, or to use a serum-free process where possible/feasible. The 9CFR test protocols were written from the perspective of heat inactivation of the serum [9CFR 113.53(c)]. Gamma irradiation of serum has become the industry standard for human biologicals as recommended for FBS used in Europe [CPMP/BWP/1793/02]. Sera are usually irradiated using a dose of 20–25kGy, while for veterinary use, the dose is recommended to be 35kGy [128]. Some viruses are relatively insensitive to gamma irradiation and it would be important to determine how 25kgray of gamma irradiation affected viruses of concern.
For trypsin
Manufacturers should also continue to be encouraged to use recombinant trypsin, as well as suspension production that minimizes trypsin use, whenever feasible; however, there will still be a legacy of serum and trypsin use for older cell lines and these should be thoroughly screened for virus contamination. It should be kept in mind, however, that the process for preparing recombinant trypsin must itself be considered for sources of viral contamination [149].
4.5. Consider incorporating the minipool concept
Commercial batches of fetal bovine serum usually consist of pooled sera collected from hundreds to >1000 animals [1]. In the human blood industry, a minipool approach is used where 6–8 units are pooled and tested prior to pooling into a larger batch for fractionation. If the minipool concept were to be incorporated for FBS, it should increase the sensitivity of the testing and help to avoid the risk of contaminating larger serum pools. One drawback to the minipool approach is that it would almost certainly increase the cost of serum, and thus, may increase the risk of black-market practices and influx of illicit serum into the industry (see discussion above about this concern).
4.6. Consider testing for specified additional viruses
Another possible approach would be to consider the necessity and merit of including specific testing for additional viruses not specifically categorized as bovine or porcine viruses (such as arboviruses), which can infect these animals. Additionally, consideration should be given to whether specific testing needs to be performed for more recently discovered viruses, e.g., hokovirus or circoviruses.
4.7. Continually monitor reports of new viruses or changes in geographic source or host range
Agents that were not believed to be of concern for cattle or swine may cross species barriers. One recent example is that of the Reston strain of Ebola virus that has been found to infect pigs in the Philippines and workers on the swine farms have seroconverted [109]. Concern also exists for agents that are not obviously productively infectious for humans or for human cells in vitro, because they may have the potential to become latent and may be oncogenic in non-host species.
4.8. Continue to consider geographic sources of raw materials
The geographic sources of raw materials are critical, not only because of BSE, but also for other potential contaminating agents. Viruses that are known to be present in Australia and New Zealand [which in addition to the U.S. are the main sources of fetal bovine serum] are important to review and consider. Geographic sourcing remains an important aspect of quality control for these raw materials.
4.9. Incorporate virus-family testing of raw materials
Grouping viruses of concern because of human host range by virus family may allow streamlining of testing of raw materials for contaminants using PCR for conserved sequences or testing for related viruses using cell lines sensitive to viruses in a particular family. As shown in Table 5, 21 virus families contain viruses with bovine host range and human host range and 17 virus families include viruses with porcine host range and human host range. Since information was often not available for each member of a family, attention should be given to groups of closely related viruses where evidence of human/porcine or human/bovine host range has been found. As a result, assays to detect closely related viruses within specific families might be of great value. Since so many bovine and porcine viruses are a potential contamination risk, addition of virus family-specific PCR assays, or other similar technology of equivalent or better sensitivity, to detect all/most members of each of the 21 virus families that have been reported to have bovine and human host range could reduce the risk of human infection. For a virus that cannot grow readily in cell culture, a PCR-based assay for the virus may be a useful direct detection method. Once viral nucleic acids have been identified by such methods, it would still be important to demonstrate whether their presence reflects an infectious agent. See Schuurman et al [150] for a bovine polyomavirus assay where virus isolation correlated with PCR detection.
4.10. Consider additional in-process testing
Manufacturers who use serum or trypsin in their cell cultures or elsewhere in their processes could add an additional test to the viral screening of their unprocessed bulks. One approach to be considered would be to include an additional bovine and/or porcine cell line, as appropriate, with the in vitro testing that is already done with three or more cell lines, generally of human and monkey origin and of the species of origin of the production cell substrate.
4.11. Encourage or require evaluation of the ability of bovine or porcine viruses to propagate in the cell substrate
Another possible enhancement to the current situation would be to encourage or require that manufacturers screen their production cell line for the ability of bovine or porcine viruses likely to be contaminants of FBS or porcine trypsin to replicate in their cells. In this way, they may focus their virus-specific testing on detection of the most likely contaminants.
4.12. Consider use of new testing technologies
Regulators and manufacturers should consider the introduction of one or more of the new testing methodologies that have been developed in the last few years for the testing of serum and trypsin. These may be useful as adjuncts and/or replacements for the 9CFR tests. Tests that are routinely used for additional testing of cell banks and end-of-production cells or unprocessed bulk samples to check for contaminating viruses include in vitro or in vivo virus screens, examination of cell sections by transmission electron microscopy (TEM), retrovirus testing (e.g., RT, PERT) and virus-specific PCR tests. New tests under development include PCR with mass spectrometry (Ibis T5000) [151], virus microarrays (VIROCHIP) [152] and massively parallel (deep) sequencing. These procedures offer promise for identification of a wider range of potential viruses and need to be carefully evaluated under field conditions for sensitivity and specificity as well as ease-of-use, time-to-results, and cost-effectiveness. As with traditional PCR, most of these methods would detect nucleic acids, without regard to whether these represent infectious viruses or not, and follow-up testing for any contaminants detected by these methods would be necessary.
5. Role of the Funding Source
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400045C. Support was also provided by Biotechnology Assessment Services, Inc.
The opinions expressed are solely those of the authors and should not be taken to represent the views or official policies of the NIH/NIAID or the U.S. government.
Abbreviations
- 9CFR
Code of Federal Regulations, Title 9
- Ab
Antibody
- BT
Bovine Turbinate cells
- CBER
Center for Biologics Evaluation and Research
- CDER
Center for Drug Evaluation and Research
- FBS
Fetal Bovine Serum
- HAd
Hemadsorption
- HHR
Human Host Range
- ICTV
International Committee on Taxonomy of Viruses
- IFA
Immunofluorescence Assay
- PCR
Polymerase Chain Reaction
- PERT
Product-Enhanced Reverse Transcriptase assay
- RBC
Red blood cells
- RT
Reverse Transcriptase
- ST
Swine Testicular cells
- TEM
Transmission Electron Microscopy
- USDA
Unites States Department of Agriculture
- VERO
African green-monkey kidney cell line
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Carol Marcus-Sekura, Email: crsek@aol.com.
James C. Richardson, Email: james.richardson@ablinc.com.
Rebecca K. Harston, Email: rebecca.harston@ablinc.com.
Nandini Sane, Email: nandini.sane@ablinc.com.
Rebecca L. Sheets, Email: RSheets@niaid.nih.gov.
6. References
- 1.Jochems CEA. Use, Trade and Harvest of Livestock Sera. Department of Laboratory Animal Science, Department of Animal Sciences, Utrecht University, Wageningen Agricultural University; 1997. p. 64. [Google Scholar]
- 2.Campbell GL, Mataczynski JD, Reisdorf ES, Powell JW, Martin DA, Lambert AJ, Haupt TE, Davis JP, Lanciotti RS. Second human case of Cache Valley virus disease. Emerg Infect Dis. 2006;12:854. doi: 10.3201/eid1205.051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bethencourt V. Virus stalls Genzyme plant. Nature Biotechnology. 2009;27:681. [Google Scholar]
- 4.Nims RW. Detection of adventitious viruses in biologicals--a rare occurrence. Dev Biol (Basel) 2006;123:153. [PubMed] [Google Scholar]
- 5.Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, Delwart EL. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol. 2010;84:6033. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.FDA. Vaccines, Blood & Biologics. 2010. FDA Actions. [Google Scholar]
- 7.M ICTVdB. 00.001.0.01.010.00.309. Bovine adenovirus 9. In: Büchen-Osmond CE, editor. ICTVdB - The Universal Virus Database, version 4. Columbia University; New York, US, New York: 2006. [Google Scholar]
- 8.Brassard J, Gagne MJ, Lamoureux L, Inglis GD, Leblanc D, Houde A. Molecular detection of bovine and porcine Torque teno virus in plasma and feces. Vet Microbiol. 2008;126:271. doi: 10.1016/j.vetmic.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 9.M ICTVdb. 01.081.0.01.001. Borna disease virus. In: Büchen-Osmond CE, editor. ICTVdB - The Universal Virus Database, version 4. Columbia University; New York, US: 2006. [Google Scholar]
- 10.Carbone KM. Borna disease virus and human disease. Clin Microbiol Rev. 2001;14:513. doi: 10.1128/CMR.14.3.513-527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode L, Ludwig H. Borna disease virus infection, a human mental-health risk. Clin Microbiol Rev. 2003;16:534. doi: 10.1128/CMR.16.3.534-545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bode L, Dietrich DE, Stoyloff R, Emrich HM, Ludwig H. Amantadine and human Borna disease virus in vitro and in vivo in an infected patient with bipolar depression. Lancet. 1997;349:178. doi: 10.1016/S0140-6736(05)60979-8. [DOI] [PubMed] [Google Scholar]
- 13.Boughton CR, Hawkes RA, Naim HM. Arbovirus infection in humans in NSW: seroprevalence and pathogenicity of certain Australian bunyaviruses. Aust N Z J Med. 1990;20:51. doi: 10.1111/j.1445-5994.1990.tb00371.x. [DOI] [PubMed] [Google Scholar]
- 14.Ishibashi K, Shirakawa H, Uchinuno Y, Ogawa T. Seroprevalence survey of Aino virus infection in dairy cattle of Fukuoka, Japan in 1990. J Vet Med Sci. 1995;57:1. doi: 10.1292/jvms.57.1. [DOI] [PubMed] [Google Scholar]
- 15.Eloit M. Risks of virus transmission associated with animal sera or substitutes and methods of control. Dev Biol Stand. 1999;99:9. [PubMed] [Google Scholar]
- 16.Pensiero MN, Sharefkin JB, Dieffenbach CW, Hay J. Hantaan virus infection of human endothelial cells. J Virol. 1992;66:5929. doi: 10.1128/jvi.66.10.5929-5936.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahr U, Muranyi W, Muller S, Kehm R, Handermann M, Darai G, Zeier M. Bovine aortic endothelial cells are susceptible to Hantaan virus infection. Virology. 2004;321:1. doi: 10.1016/j.virol.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Danes L, Pejcoch M, Bukovjan K, Veleba J, Halackova M. [Antibodies against Hantaviruses in game and domestic oxen in the Czech Republic] Cesk Epidemiol Mikrobiol Imunol. 1992;41:15. [PubMed] [Google Scholar]
- 19.Godsey MS, Jr, Amoo F, Yuill TM, Defoliart GR. California serogroup virus infections in Wisconsin domestic animals. Am J Trop Med Hyg. 1988;39:409. doi: 10.4269/ajtmh.1988.39.409. [DOI] [PubMed] [Google Scholar]
- 20.Grimstad PR, Artsob H, Karabatsos N, Calisher CH. Production and use of a hemagglutinin for detecting antibody to Jamestown Canyon virus. J Clin Microbiol. 1987;25:1557. doi: 10.1128/jcm.25.8.1557-1559.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerhardt RR, Gottfried KL, Apperson CS, Davis BS, Erwin PC, Smith AB, Panella NA, Powell EE, Nasci RS. First isolation of La Crosse virus from naturally infected Aedes albopictus. Emerg Infect Dis. 2001;7:807. doi: 10.3201/eid0705.017506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CFSPH. Hantavirus. The Center for Food Security and Public Health, Iowa State University; Ames, Iowa: 2008. [Google Scholar]
- 23.OIE. OIE. The World Organization for Animal Health (OIE); Paris, France: 2009. [Google Scholar]
- 24.Mattison K, Shukla A, Cook A, Pollari F, Friendship R, Kelton D, Bidawid S, Farber JM. Human noroviruses in swine and cattle. Emerg Infect Dis. 2007;13:1184. doi: 10.3201/eid1308.070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Der Poel WH, Vinje J, van Der Heide R, Herrera MI, Vivo A, Koopmans MP. Norwalk-like calicivirus genes in farm animals. Emerg Infect Dis. 2000;6:36. doi: 10.3201/eid0601.000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AW, Skilling DE, Cherry N, Mead JH, Matson DO. Calicivirus emergence from ocean reservoirs: zoonotic and interspecies movements. Emerg Infect Dis. 1998;4:13. doi: 10.3201/eid0401.980103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hattermann K, Roedner C, Schmitt C, Finsterbusch T, Steinfeldt T, Mankertz A. Infection studies on human cell lines with porcine circovirus type 1 and porcine circovirus type 2. Xenotransplantation. 2004;11:284. doi: 10.1111/j.1399-3089.2004.00134.x. [DOI] [PubMed] [Google Scholar]
- 28.Nayar GP, Hamel AL, Lin L, Sachvie C, Grudeski E, Spearman G. Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can Vet J. 1999;40:277. [PMC free article] [PubMed] [Google Scholar]
- 29.Alekseev KP, Vlasova AN, Jung K, Hasoksuz M, Zhang X, Halpin R, Wang S, Ghedin E, Spiro D, Saif LJ. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J Virol. 2008;82:12422. doi: 10.1128/JVI.01586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smits SL, Lavazza A, Matiz K, Horzinek MC, Koopmans MP, de Groot RJ. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J Virol. 2003;77:9567. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwabara M, Wada K, Maeda Y, Miyazaki A, Tsunemitsu H. First isolation of cytopathogenic bovine torovirus in cell culture from a calf with diarrhea. Clin Vaccine Immunol. 2007;14:998. doi: 10.1128/CVI.00475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Audet SA, Crim RL, Beeler J. Evaluation of vaccines, interferons and cell substrates for pestivirus contamination. Biologicals. 2000;28:41. doi: 10.1006/biol.1999.0240. [DOI] [PubMed] [Google Scholar]
- 33.Kahrs R. Viral Diseases of Cattle. Iowa University Press; Ames: 2001. [Google Scholar]
- 34.Beran G. Section B: Viral Zoonoses. In: Steele JH, Beran G, editors. Handbook Of Zoonoses. CRC Press; Boca Raton, Florida: 1994. p. 582. [Google Scholar]
- 35.CFSPH. Louping ill. The Center for Food Security and Public Health, Iowa State University; 2009. [Google Scholar]
- 36.Ulloa A, Langevin SA, Mendez-Sanchez JD, Arredondo-Jimenez JI, Raetz JL, Powers AM, Villarreal-Trevino C, Gubler DJ, Komar N. Serologic survey of domestic animals for zoonotic arbovirus infections in the Lacandon Forest region of Chiapas, Mexico. Vector Borne Zoonotic Dis. 2003;3:3. doi: 10.1089/153036603765627406. [DOI] [PubMed] [Google Scholar]
- 37.Sikutova S, Hornok S, Hubalek Z, Dolezalkova I, Juricova Z, Rudolf I. Serological survey of domestic animals for tick-borne encephalitis and Bhanja viruses in northeastern Hungary. Vet Microbiol. 2009;135:267. doi: 10.1016/j.vetmic.2008.09.082. [DOI] [PubMed] [Google Scholar]
- 38.Ozkul A, Yildirim Y, Pinar D, Akcali A, Yilmaz V, Colak D. Serological evidence of West Nile Virus (WNV) in mammalian species in Turkey. Epidemiol Infect. 2006;134:826. doi: 10.1017/S0950268805005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banks M, Bendall R, Grierson S, Heath G, Mitchell J, Dalton H. Human and porcine hepatitis E virus strains, United Kingdom. Emerg Infect Dis. 2004;10:953. doi: 10.3201/eid1005.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper K, Huang FF, Batista L, Rayo CD, Bezanilla JC, Toth TE, Meng XJ. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol. 2005;43:1684. doi: 10.1128/JCM.43.4.1684-1688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng Y, Wang C, Zhao C, Yu X, Harrison TJ, Tian K, Wang Y. Serological prevalence of hepatitis E virus in domestic animals and diversity of genotype 4 hepatitis E virus in China. Vector Borne Zoonotic Dis. 2010;10:765. doi: 10.1089/vbz.2009.0168. [DOI] [PubMed] [Google Scholar]
- 42.Gillet L, Minner F, Detry B, Farnir F, Willems L, Lambot M, Thiry E, Pastoret PP, Schynts F, Vanderplasschen A. Investigation of the susceptibility of human cell lines to bovine herpesvirus 4 infection: demonstration that human cells can support a nonpermissive persistent infection which protects them against tumor necrosis factor alpha-induced apoptosis. J Virol. 2004;78:2336. doi: 10.1128/JVI.78.5.2336-2347.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machiels B, Gillet L, Nascimento Brito SD, Drion P, Delforge C, Nizet Y, Gianello P, Bona C, Costes B, Markine-Goriaynoff N, Vanderplasschen A. Natural antibody--complement dependent neutralization of bovine herpesvirus 4 by human serum. Microbes Infect. 2007;9:1530. doi: 10.1016/j.micinf.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Roumillat LF, Feorino PM, Lukert PD. Persistent infection of a human lymphoblastoid cell line with equine herpesvirus 1. Infect Immun. 1979;24:539. doi: 10.1128/iai.24.2.539-544.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crandell RA, Ichimura H, Kit S. Isolation and comparative restriction endonuclease DNA fingerprinting of equine herpesvirus-1 from cattle. Am J Vet Res. 1988;49:1807. [PubMed] [Google Scholar]
- 46.Michalski FJ, Dietz A, Hsiung GD. Growth characteristics of bovine herpesvirus 1 (infectious bovine rhinotracheitis) in human diploid cell strain WI-38. Proc Soc Exp Biol Med. 1976;151:407. doi: 10.3181/00379727-151-39221. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi S, Ogawa S, Takashima Y, Otsuka H. The neutralization of pseudorabies virus by anti-alpha-galactocyl natural antibody in normal serum. Virus Res. 2004;99:1. doi: 10.1016/j.virusres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Jones TC, Hunt RD, King NW. In: Veterinary Pathology. Jones HRTC, King NW, editors. Wiley-Blackwell; Baltimore: 1997. p. 1392. [Google Scholar]
- 49.M ICTVdb. 00.046.0.03. Thogotovirus. In: Büchen-Osmond CE, editor. ICTVdB - The Universal Virus Database, version 4. Columbia University; New York, US: 2006. [Google Scholar]
- 50.Gunning RF, Brown IH, Crawshaw TR. Evidence of influenza A virus infection in dairy cows with sporadic milk drop syndrome. Vet Rec. 1999;145:556. doi: 10.1136/vr.145.19.556. [DOI] [PubMed] [Google Scholar]
- 51.Kalthoff D, Hoffmann B, Harder T, Durban M, Beer M. Experimental infection of cattle with highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:1132. doi: 10.3201/eid1407.071468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore DL, Causey OR, Carey DE, Reddy S, Cooke AR, Akinkugbe FM, David-West TS, Kemp GE. Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann Trop Med Parasitol. 1975;69:49. doi: 10.1080/00034983.1975.11686983. [DOI] [PubMed] [Google Scholar]
- 53.Bonnez W, Reichman RC, Strussenberg J, Roberts NJ., Jr In vitro interactions between bovine papillomavirus and human monocytes and macrophages. Intervirology. 1991;32:246. doi: 10.1159/000150206. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Ishai Z, Naftali V, Avram A, Yatziv S. Human infection by a bovine strain of parainfluenza virus type 3. J Med Virol. 1980;6:165. doi: 10.1002/jmv.1890060209. [DOI] [PubMed] [Google Scholar]
- 55.Haller AA, Mitiku M, MacPhail M. Bovine parainfluenza virus type 3 (PIV3) expressing the respiratory syncytial virus (RSV) attachment and fusion proteins protects hamsters from challenge with human PIV3 and RSV. J Gen Virol. 2003;84:2153. doi: 10.1099/vir.0.19079-0. [DOI] [PubMed] [Google Scholar]
- 56.Schlender J, Zimmer G, Herrler G, Conzelmann KK. Respiratory syncytial virus (RSV) fusion protein subunit F2, not attachment protein G, determines the specificity of RSV infection. J Virol. 2003;77:4609. doi: 10.1128/JVI.77.8.4609-4616.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva AC, Delgado I, Sousa MF, Carrondo MJ, Alves PM. Scalable culture systems using different cell lines for the production of Peste des Petits ruminants vaccine. Vaccine. 2008;26:3305. doi: 10.1016/j.vaccine.2008.03.077. [DOI] [PubMed] [Google Scholar]
- 58.Farooq U, Khan QM, Barrett T. Molecular based diagnosis of rinderpest and peste des petits ruminants virus in Pakistan. Int J Agri Biol. 2008;10:93. [Google Scholar]
- 59.Liess B, Plowright W. The Propagation and Growth Characteristics of Rinderpest Virus in Hela Cells. Arch Gesamte Virusforsch. 1963;14:27. doi: 10.1007/BF01555160. [DOI] [PubMed] [Google Scholar]
- 60.Baron MD, Barrett T. Rescue of rinderpest virus from cloned cDNA. J Virol. 1997;71:1265. doi: 10.1128/jvi.71.2.1265-1271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt M, Katano H, Bossis I, Chiorini JA. Cloning and characterization of a bovine adeno-associated virus. J Virol. 2004;78:6509. doi: 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lau SK, Woo PC, Tse H, Fu CT, Au WK, Chen XC, Tsoi HW, Tsang TH, Chan JS, Tsang DN, Li KS, Tse CW, Ng TK, Tsang OT, Zheng BJ, Tam S, Chan KH, Zhou B, Yuen KY. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J Gen Virol. 2008;89:1840. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- 63.Smyth M, Symonds A, Brazinova S, Martin J. Bovine enterovirus as an oncolytic virus: foetal calf serum facilitates its infection of human cells. Int J Mol Med. 2002;10:49. doi: 10.3892/ijmm.10.1.49. [DOI] [PubMed] [Google Scholar]
- 64.Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84:3069. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]
- 65.Khamrin P, Okitsu S, Ushijima H, Maneekarn N. Novel nonstructural protein 4 genetic group in rotavirus of porcine origin. Emerg Infect Dis. 2008;14:686. doi: 10.3201/eid1404.07111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberste MS, Gotuzzo E, Blair P, Nix WA, Ksiazek TG, Comer JA, Rollin P, Goldsmith CS, Olson J, Kochel TJ. Human febrile illness caused by encephalomyocarditis virus infection, Peru. Emerg Infect Dis. 2009;15:640. doi: 10.3201/eid1504.081428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomson GR, Bengis RG, Brown CC. In: Infectious Diseases of Wild Mammals. Williams ES, Barker IK, editors. Iowa State University Press; Ames: 2001. [Google Scholar]
- 68.Aftosa F. Foot and Mouth Disease. CFSPH, The Center for Food Security and Public Health, Iowa State University; Ames: 2007. [Google Scholar]
- 69.Prempeh H, Smith R, Muller B. Foot and mouth disease: the human consequences. The health consequences are slight, the economic ones huge. Bmj. 2001;322:565. doi: 10.1136/bmj.322.7286.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hales LM, Knowles NJ, Reddy PS, Xu L, Hay C, Hallenbeck PL. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J Gen Virol. 2008;89:1265. doi: 10.1099/vir.0.83570-0. [DOI] [PubMed] [Google Scholar]
- 71.Parry JV, Gardner SD. Human exposure to bovine polyomavirus: a zoonosis? Arch Virol. 1986;87:287. doi: 10.1007/BF01315306. [DOI] [PubMed] [Google Scholar]
- 72.de Souza Trindade G, da Fonseca FG, Marques JT, Nogueira ML, Mendes LC, Borges AS, Peiro JR, Pituco EM, Bonjardim CA, Ferreira PC, Kroon EG. Aracatuba virus: a vaccinialike virus associated with infection in humans and cattle. Emerg Infect Dis. 2003;9:155. doi: 10.3201/eid0902.020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schnurrenberger PR, Swango LJ, Bowman GM, Luttgen PJ. Bovine papular stomatitis incidence in veterinary students. Can J Comp Med. 1980;44:239. [PMC free article] [PubMed] [Google Scholar]
- 74.Damaso CR, Esposito JJ, Condit RC, Moussatche N. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology. 2000;277:439. doi: 10.1006/viro.2000.0603. [DOI] [PubMed] [Google Scholar]
- 75.NADIS: TheCattleSite: Diseases and Conditions. National Animal Disease Information Service, LTD; Sheffield, England: [Google Scholar]
- 76.Damon IK. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott Williams and Wilkins; Philadelphia: 2006. [Google Scholar]
- 77.Nabeshima T, Thi Nga P, Guillermo P, Parquet Mdel C, Yu F, Thanh Thuy N, Minh Trang B, Tran Hien N, Sinh Nam V, Inoue S, Hasebe F, Morita K. Isolation and molecular characterization of Banna virus from mosquitoes, Vietnam. Emerg Infect Dis. 2008;14:1276. doi: 10.3201/eid1408.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohd Jaafar F, Attoui H, Mertens PP, de Micco P, de Lamballerie X. Identification and functional analysis of VP3, the guanylyltransferase of Banna virus (genus Seadornavirus, family Reoviridae) J Gen Virol. 2005;86:1141. doi: 10.1099/vir.0.80579-0. [DOI] [PubMed] [Google Scholar]
- 79.CFSPH. Bluetongue: Sore Muzzle, Pseudo Foot-and-Mouth Disease, Muzzle Disease. Iowa State University, the Center for Food Security and Public Health; Ames, Iowa: 2006. [Google Scholar]
- 80.Mettler NE, Macnamara LG, Shope RE. The propagation of the virus of epizootic hemorrhagic disease of deer in newborn mice and HeLa cells. J Exp Med. 1962;116:665. doi: 10.1084/jem.116.5.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.CFSPH. Diseases caused by the Epizootic Hemorrhagic Disease Virus Serogroup. The Center for Food Security and Public Health, Iowa State University; 2006. [Google Scholar]
- 82.Attoui H, Mohd Jaafar F, Belhouchet M, Tao S, Chen B, Liang G, Tesh RB, de Micco P, de Lamballerie X. Liao ning virus, a new Chinese seadornavirus that replicates in transformed and embryonic mammalian cells. J Gen Virol. 2006;87:199. doi: 10.1099/vir.0.81294-0. [DOI] [PubMed] [Google Scholar]
- 83.Moore WA. Experience in cell line testing. Dev Biol Stand. 1992;76:51. [PubMed] [Google Scholar]
- 84.Bailey K. UPDATE LETTER TO HOSPITALS RE ABBOKINASE (UROKINASE) Health Canada; 1999. [Google Scholar]
- 85.M ICTVdb. 00.060.0.03. Rotavirus. In: Büchen-Osmond CE, editor. ICTVdB - The Universal Virus Database, version 4. Columbia University; New York, US: 2006. [Google Scholar]
- 86.Terrett LA, Saif LJ, Theil KW, Kohler EM. Physicochemical characterization of porcine pararotavirus and detection of virus and viral antibodies using cell culture immunofluorescence. J Clin Microbiol. 1987;25:268. doi: 10.1128/jcm.25.2.268-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma Z, Hao P, Yao X, Liu C, Tan J, Liu L, Yang R, Geng Y, Chen Q, Qiao W. Establishment of an indicator cell line to quantify bovine foamy virus infection. J Basic Microbiol. 2008;48:278. doi: 10.1002/jobm.200700295. [DOI] [PubMed] [Google Scholar]
- 88.Buehring GC, Philpott SM, Choi KY. Humans have antibodies reactive with Bovine leukemia virus. AIDS Res Hum Retroviruses. 2003;19:1105. doi: 10.1089/088922203771881202. [DOI] [PubMed] [Google Scholar]
- 89.Bethwaite P, McLean D, Kennedy J, Pearce N. Adult-onset acute leukemia and employment in the meat industry: a New Zealand case-control study. Cancer Causes Control. 2001;12:635. doi: 10.1023/a:1011203809049. [DOI] [PubMed] [Google Scholar]
- 90.Pearce N, Smith AH, Reif JS. Increased risks of soft tissue sarcoma, malignant lymphoma, and acute myeloid leukemia in abattoir workers. Am J Ind Med. 1988;14:63. doi: 10.1002/ajim.4700140108. [DOI] [PubMed] [Google Scholar]
- 91.Hirsh YCDCZ. Veterinary Microbiology. Blackwell Science; 1999. p. 479. [Google Scholar]
- 92.WHO. Global Early Warning System for Major Animal Diseases, including Zoonoses (GLEWS): Triggers and GLEWS diseases. World Health Oranization; 2009. WHO. [Google Scholar]
- 93.5M. ThePigSite Quick Disease Guide. 5M Enterprises Ltd; Sheffield, England: 2009. Vesicular Stomatitis. [Google Scholar]
- 94.LCP Health: Veterinary Public Health. Overview of Zoonoses. COUNTY OF LOS ANGELES - DEPARTMENT OF HEALTH SERVICES; Los Angeles, CA: [Google Scholar]
- 95.McGee ED, Littleton CH, Mapp JB, Brown RJ. Eastern equine encephalomyelitis in an adult cow. Vet Pathol. 1992;29:361. doi: 10.1177/030098589202900414. [DOI] [PubMed] [Google Scholar]
- 96.Scott TW. In: The Arboviruses: Epidemiology and Ecology. Monath TP, editor. CRC Press; Boca Raton: 1988. [Google Scholar]
- 97.Sanderson CJ. A serologic survey of Queensland cattle for evidence of arbovirus infections. Am J Trop Med Hyg. 1969;18:433. doi: 10.4269/ajtmh.1969.18.433. [DOI] [PubMed] [Google Scholar]
- 98.CFSPH. Getah Virus Infection. The Center for Food Security and Public Health, Iowa State University; Ames: 2006. [Google Scholar]
- 99.Harley D, Sleigh A, Ritchie S. Ross River virus transmission, infection, and disease: a cross-disciplinary review. Clin Microbiol Rev. 2001;14:909. doi: 10.1128/CMR.14.4.909-932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Navarro JC, Medina G, Vasquez C, Coffey LL, Wang E, Suarez A, Biord H, Salas M, Weaver SC. Postepizootic persistence of Venezuelan equine encephalitis virus, Venezuela. Emerg Infect Dis. 2005;11:1907. doi: 10.3201/eid1112.050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Griffin DE. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott Williams and Wilkins; Philadelphia: 2006. [Google Scholar]
- 102.Hansman GS, Oka T, Okamoto R, Nishida T, Toda S, Noda M, Sano D, Ueki Y, Imai T, Omura T, Nishio O, Kimura H, Takeda N. Human sapovirus in clams, Japan. Emerg Infect Dis. 2007;13:620. doi: 10.3201/eid1304.061390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tischer I, Bode L, Apodaca J, Timm H, Peters D, Rasch R, Pociuli S, Gerike E. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch Virol. 1995;140:1427. doi: 10.1007/BF01322669. [DOI] [PubMed] [Google Scholar]
- 104.Storz J, Zhang XM, Rott R. Comparison of hemagglutinating, receptor-destroying, and acetylesterase activities of avirulent and virulent bovine coronavirus strains. Arch Virol. 1992;125:193. doi: 10.1007/BF01309637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Welter MW. Adaptation and serial passage of bovine coronavirus in an established diploid swine testicular cell line and subsequent development of a modified live vaccine. Adv Exp Med Biol. 1998;440:707. doi: 10.1007/978-1-4615-5331-1_91. [DOI] [PubMed] [Google Scholar]
- 106.Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weingartl HM, Copps J, Drebot MA, Marszal P, Smith G, Gren J, Andova M, Pasick J, Kitching P, Czub M. Susceptibility of pigs and chickens to SARS coronavirus. Emerg Infect Dis. 2004;10:179. doi: 10.3201/eid1002.030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Terao Y, Takagi H, Phan TG, Okitsu S, Ushijima H. Identification of antibody against porcine coronavirus in human milk. Clin Lab. 2007;53:129. [PubMed] [Google Scholar]
- 109.WHO. Ebola Reston in pigs and humans in the Philippines. WHO; Geneva, Switzerland: 2009. Ebola Reston in pigs and humans in the Philippines. [Google Scholar]
- 110.THSS Burgess, MD, US), Porter, Kevin R. (Boyds, MD, US), Freilich, Daniel A. (Washington, DC, US), Doolan, Denise L. (Camp Hill, AU) (2008): Method for the evaluation of dengue virus therapeutic agents, United States.
- 111.Leyssen P, De Clercq E, Neyts J. Perspectives for the treatment of infections with Flaviviridae. Clin Microbiol Rev. 2000;13:67. doi: 10.1128/cmr.13.1.67-82.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McLean DM, Smith PA, Livingstone SE, Wilson WE, Wilson AG. Powassan virus: vernal spread during 1965. Can Med Assoc J. 1966;94:532. [PMC free article] [PubMed] [Google Scholar]
- 113.Bugrysheva JV, Matveeva VA, Dobrikova EY, Bykovskaya NV, Korobova SA, Bakhvalova VN, Morozova OV. Tick-borne encephalitis virus NS1 glycoprotein during acute and persistent infection of cells. Virus Res. 2001;76:161. doi: 10.1016/s0168-1702(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 114.Paranjape S, Patil BR, Kadam VD. Characterization of porcine stable kidney cell line adapted to hyperthermic temperature. In Vitro Cell Dev Biol Anim. 2003;39:193. doi: 10.1290/1543-706X(2003)039<0193:COPSKC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 115.Scheidler A, Rokos K, Reuter T, Ebermann R, Pauli G. Inactivation of viruses by beta-propiolactone in human cryo poor plasma and IgG concentrates. Biologicals. 1998;26:135. doi: 10.1006/biol.1998.0125. [DOI] [PubMed] [Google Scholar]
- 116.Van Reeth K. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet Res. 2007;38:243. doi: 10.1051/vetres:2006062. [DOI] [PubMed] [Google Scholar]
- 117.Potts BJ, Kivens WJ. Detecting and removing porcine viruses. BioPharm International; 2000. [Google Scholar]
- 118.CFSPH. Menangle. The Center for Food Security and Public Health, Iowa State University; 2008. [Google Scholar]
- 119.Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, Daniels PW, Gould AR, Hyatt AD. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg Infect Dis. 1998;4:269. doi: 10.3201/eid0402.980214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bowden TR, Westenberg M, Wang LF, Eaton BT, Boyle DB. Molecular characterization of Menangle virus, a novel paramyxovirus which infects pigs, fruit bats, and humans. Virology. 2001;283:358. doi: 10.1006/viro.2001.0893. [DOI] [PubMed] [Google Scholar]
- 121.Bowden TA, Crispin M, Harvey DJ, Aricescu AR, Grimes JM, Jones EY, Stuart DI. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82:11628. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luby SP, Rahman M, Hossain MJ, Blum LS, Husain MM, Gurley E, Khan R, Ahmed BN, Rahman S, Nahar N, Kenah E, Comer JA, Ksiazek TG. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12:1888. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goh KJ, Tan CT, Chew NK, Tan PS, Kamarulzaman A, Sarji SA, Wong KT, Abdullah BJ, Chua KB, Lam SK. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342:1229. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- 125.Nawathe DTWP. Experimental infection of domestic pigs with the virus of peste des petits ruminants. Trop Anim Health Prod. 1979;11:120. doi: 10.1007/BF02237785. [DOI] [PubMed] [Google Scholar]
- 126.CFSPH. Rinderpest. The Center for Food Security and Public Health, Iowa State University; Ames: 2008. [Google Scholar]
- 127.Yaiw KC, Bingham J, Crameri G, Mungall B, Hyatt A, Yu M, Eaton B, Shamala D, Wang LF, Thong Wong K. Tioman virus, a paramyxovirus of bat origin, causes mild disease in pigs and has a predilection for lymphoid tissues. J Virol. 2008;82:565. doi: 10.1128/JVI.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Merten OW. Virus contaminations of cell cultures - A biotechnological view. Cytotechnology. 2002;39:91. doi: 10.1023/A:1022969101804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Knowles NJ, Buckley LS, Pereira HG. Classification of porcine enteroviruses by antigenic analysis and cytopathic effects in tissue culture: description of 3 new serotypes. Arch Virol. 1979;62:201. doi: 10.1007/BF01317552. [DOI] [PubMed] [Google Scholar]
- 130.AFFA. Generic Import Risk Analysis (IRA) for Uncooked Pig Meat. 2001. p. 105. [Google Scholar]
- 131.Specke V, Rubant S, Denner J. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology. 2001;285:177. doi: 10.1006/viro.2001.0934. [DOI] [PubMed] [Google Scholar]
- 132.Elvinger F, Baldwin CA, Liggett AD, Tang KN, Stallknecht DE. Prevalence of exposure to eastern equine encephalomyelitis virus in domestic and feral swine in Georgia. J Vet Diagn Invest. 1996;8:481. doi: 10.1177/104063879600800414. [DOI] [PubMed] [Google Scholar]
- 133.Brown C, Torres A, editors. CoFADotUSAHA USAHA. Foreign Animal Diseases. 2008. [Google Scholar]
- 134.Pauli G, Grunmach J, Ludwig H. Focus-immunoassay for Borna disease virus-specific antigens. Zentralbl Veterinarmed B. 1984;31:552. doi: 10.1111/j.1439-0450.1984.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 135.Horling J, Lundkvist A, Persson K, Mullaart M, Dzagurova T, Dekonenko A, Tkachenko E, Niklasson B. Detection and subsequent sequencing of Puumala virus from human specimens by PCR. J Clin Microbiol. 1995;33:277. doi: 10.1128/jcm.33.2.277-282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Straub TM, Honer zu Bentrup K, Orosz-Coghlan P, Dohnalkova A, Mayer BK, Bartholomew RA, Valdez CO, Bruckner-Lea CJ, Gerba CP, Abbaszadegan M, Nickerson CA. In vitro cell culture infectivity assay for human noroviruses. Emerg Infect Dis. 2007;13:396. doi: 10.3201/eid1303.060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.CFSPH. Wesselsbron. The Center for Food Security and Public Health, Iowa State University; Ames: 2006. [Google Scholar]
- 138.Reddy PS, Burroughs KD, Hales LM, Ganesh S, Jones BH, Idamakanti N, Hay C, Li SS, Skele KL, Vasko AJ, Yang J, Watkins DN, Rudin CM, Hallenbeck PL. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J Natl Cancer Inst. 2007;99:1623. doi: 10.1093/jnci/djm198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nairn C, Lovatt A, Galbraith DN. Detection of infectious bovine polyomavirus. Biologicals. 2003;31:303. doi: 10.1016/j.biologicals.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 140.Martina BE, van Doornum G, Dorrestein GM, Niesters HG, Stittelaar KJ, Wolters MA, van Bolhuis HG, Osterhaus AD. Cowpox virus transmission from rats to monkeys, the Netherlands. Emerg Infect Dis. 2006;12:1005. doi: 10.3201/eid1206.051513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Castrucci G, McKercher DG, Cilli V, Arancia G, Azioli C. Characteristics of a paravaccinia virus from cattle. Arch Gesamte Virusforsch. 1970;29:315. doi: 10.1007/BF01249886. [DOI] [PubMed] [Google Scholar]
- 142.Cambier JC, Meek ES. Quantitative assay of paravaccinia virus based on enumeration of inclusion-containing cells. Appl Microbiol. 1972;24:138. doi: 10.1128/am.24.1.138-142.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Temizel EM, Yesilbag K, Batten C, Senturk S, Maan NS, Clement-Mertens PP, Batmaz H. Epizootic hemorrhagic disease in cattle, Western Turkey. Emerg Infect Dis. 2009;15:317. doi: 10.3201/eid1502.080572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McClurkin AW. Probable role of viruses in calfhood diseases. J Dairy Sci. 1977;60:278. doi: 10.3168/jds.S0022-0302(77)83865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Romen F, Backes P, Materniak M, Sting R, Vahlenkamp TW, Riebe R, Pawlita M, Kuzmak J, Lochelt M. Serological detection systems for identification of cows shedding bovine foamy virus via milk. Virology. 2007;364:123. doi: 10.1016/j.virol.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 146.Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 147.Abdelwahab KS, Almeida JD, Doane FW, McLean DM. Powassan Virus: Morphology and Cytopathology. Can Med Assoc J. 1964;90:1068. [PMC free article] [PubMed] [Google Scholar]
- 148.Wessman S. Vaccine Cell Substrates: Bovine and Porcine Virus Considerations. NIAID, NIH; Rockville, MD: 2004. [PubMed] [Google Scholar]
- 149.Roche AS. Trypsin, recombinant, Proteomics Grade Package insert, Version 1. Roche Diagnostics; 2003. [Google Scholar]
- 150.Schuurman R, van Steenis B, van Strien A, van der Noordaa J, Sol C. Frequent detection of bovine polyomavirus in commercial batches of calf serum by using the polymerase chain reaction. J Gen Virol. 1991;72(Pt 11):2739. doi: 10.1099/0022-1317-72-11-2739. [DOI] [PubMed] [Google Scholar]
- 151.Ecker DJ, Sampath R, Massire C, Blyn LB, Hall TA, Eshoo MW, Hofstadler SA. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol. 2008;6:553. doi: 10.1038/nrmicro1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang D. Viral Detection and Discovery using DNA Microarrays. APHL Infectious Disease Conference; 2005. p. 55. [Google Scholar]
- 153.Nakamura S, Yang CS, Sakon N, Ueda M, Tougan T, Yamashita A, Goto N, Takahashi K, Yasunaga T, Ikuta K, Mizutani T, Okamoto Y, Tagami M, Morita R, Maeda N, Kawai J, Hayashizaki Y, Nagai Y, Horii T, Iida T, Nakaya T. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Onions D, Kolman J. Massively parallel sequencing, a new method for detecting adventitious agents. Biologicals. 2010;38:377. doi: 10.1016/j.biologicals.2010.01.003. [DOI] [PubMed] [Google Scholar]