Abstract
Recent studies in mice and human identified osteocalcin (OCN) as a bone-derived hormone that modulates insulin secretion and insulin sensitivity. OCN is synthesized by the bone gamma-carboxyglutamate protein (BGLAP) gene located in the well replicated region of type 2 diabetes (T2D) linkage on chromosome 1q22. We resequenced BGLAP gene in 192 individuals with T2D and performed case-control studies in 766 Caucasian (461 T2D and 305 controls) and 563 African American individuals (371 T2D and 192 controls). Metabolic effects of BGLAP variants were examined in 127 nondiabetic members of Caucasian T2D families and in 498 unrelated nondiabetic African American and Caucasian individuals. BGLAP expression was tested in transformed lymphocytes from 60 Caucasian individuals. We identified 17 single nucleotide polymorphisms (SNPs) in African Americans, but observed only the two known SNPs in Caucasians. No SNP was associated with T2D. Promoter SNP rs1800247 was not associated with metabolic traits including insulin sensitivity (SI) or fasting glucose in either population, but nonsynonymous SNP rs34702397 (R94Q) was nominally associated with SI (uncorrected p=0.05) and glucose-mediated glucose disposal (SG; uncorrected p=0.03) in African Americans. No SNP altered measures of insulin secretion or obesity, nor was BGLAP expression associated with rs1800247. Our study was sufficiently powered to exclude BGLAP variants as a major risk factor (OR>1.5) for T2D in Caucasians, but coding variants in exon 4 may alter glucose homeostasis and diabetes risk in African Americans.
Keywords: Osteocalcin, Diabetes, Polymorphism, Transcript, Association, Insulin sensitivity
Introduction
Type 2 Diabetes (T2D) is one of the complex metabolic diseases for which convincing evidence, including twin and family studies, supports the role of genetic susceptibility loci 1. In last two years over 12 published genome wide association (GWA) scans for T2D identified at least 19 novel common variants that increase the susceptibility to T2D [14,22–23, 25]. However, with the exception of TCF7L2 the effect size of these variants is small, and together known risk variants for T2D are estimated to explain less than 5% of the inherited contribution to T2D risk [16]. Hence, many T2D risk loci likely remain to be discovered.
Recent studies in mice identified osteocalcin (OCN) as a bone-derived hormone that regulated glucose metabolism by modulating insulin secretion and insulin sensitivity [8,13]. OCN was shown to induce β-cell proliferation by stimulating CyclinD1 expression. Additionally, OCN stimulated insulin gene expression in pancreatic β-cells and adiponectin expression from adipocytes. Circulating levels of both total and the non-carboxylated active form of OCN were correlated with insulin sensitivity and insulin secretion in humans [9, 19]. OCN is synthesized by bone gamma-carboxyglutamate protein gene (BGLAP) on chromosome 1q22 in a well replicated region of linkage to T2D [4], and thus is both a functional and positional candidate for T2D susceptibility. Surprisingly, the gene is not tagged in either 1q fine mapping studies [20] or in publicly available GWA data [22–23], and thus has not been evaluated. We hypothesized that sequence polymorphisms in BGLAP or its regulatory region would alter its function or expression and contribute to T2D susceptibility by altering insulin secretion and insulin sensitivity. We sequenced BGLAP and its putative promoter region in 192 T2DM individuals of Caucasian and African American ancestry, evaluated the identified SNPs for association with T2D, and examined the effects of BGLAP SNPs on metabolic traits in Caucasian and African American populations. Furthermore, we evaluated a putative functional promoter variant for effects on transcript levels in transformed lymphocytes.
Methods
Study subjects
Case-control studies with T2D were conducted in two cohorts: a Caucasian (European American) cohort of 461 individuals with T2D and 305 nondiabetic control individuals, and in an African American cohort of 371 individuals with T2D and 192 nondiabetic control individuals. A summary of our study cohorts is provided in Supplementary Data, Table 1S. Caucasian individuals were ascertained from Utah and Arkansas (USA) for Northern European ancestry, and African American individuals were ascertained in Arkansas as described previously [3]. Individuals with T2D were on pharmacotherapy or had documented T2D by glucose tolerance tests and had at least one diabetic first degree relative. Control individuals had no family history of diabetes in a first degree relative and a normal fasting or post-challenge glucose [3]. Power to detect an association with T2D was ~80% for an OR of 1.5 at minor allele frequencies over 0.15 for both Caucasian and African American cohorts.
BGLAP SNP effects on metabolic parameters were evaluated in nondiabetic individuals of European descent from Utah (127 members of 26 families ascertained for multiple individuals with T2D) [6], and from unrelated individuals of European (n=341) or African-American (n=157) heritage ascertained in Arkansas (Supplementary Data, Table 1S). The Utah cohort underwent a tolbutamide – modified, frequently sampled intravenous glucose tolerance test (FSIGT). Because tolbutamide became unavailable part way through the study, 122 Caucasian and 69 African American subjects from Arkansas underwent tolbutamide modified FSIGT, whereas the remainder had an insulin modified FSIGT as described elsewhere [3]. All subjects provided written, informed consent under protocols approved by the Institutional Review Boards of the University of Utah Health Sciences Center, the University of Arkansas for Medical Sciences, or the Central Arkansas Veterans Healthcare System.
DNA sequencing
We sequenced a 2000 bp region that included all exons, introns, 700 bp of 5’ flanking region, and the 3’ flanking region of the BGLAP gene (chromosome 1; 154477831-154479830 bp; NCBI Build 36.1) in 7 overlapping fragments. We evaluated 96 Caucasian and 96 African American individuals with T2D. Sequencing was performed by Polymorphic DNA Technologies Inc., (Alameda, CA, USA). To ensure accuracy and sensitivity, only data from the high resolution, high quality region of the DNA sequencing runs were used. SNPs were called only when confirmed on both strands.
Genotyping
We genotyped SNPs that were polymorphic in both Caucasian and African American populations and non-synonymous SNPs identified from the dbSNP database, as well as any newly identified SNPs within the gene that showed a minor allele frequency over 0.05 in our sequenced samples. SNPs were genotyped by Pyrosequencing (PSQ96, Qiagen Inc, Valencia, CA, USA), and typing confirmed by 100% concordance in 10% blinded duplicates.
Gene expression
Total RNA from transformed lymphocytes grown under standard conditions was extracted using RNEasy mini kit (Qiagene Inc) and reverse transcribed using random hexamers (Taqman Reverse Transcription Reagents, Applied Biosystems, Inc, Foster City, CA). BGLAP expression was measured by quantitative real time PCR using SYBR green chemistry (Applied Biosystems, Inc.) and normalized to 18S ribosomal RNA. Primers excluded the overlapping PMF1 gene and were designed for exon 1 and junction of exons 1 and 2, are available from the authors.
Statistical analysis
Comparison of allele frequencies in case-control cohorts were performed using Fisher Exact and Cochran-Armitage Trend tests. Hardy-Weinberg equilibrium was tested by using exact test in the online DeFinetti program (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl). Linkage disequilibrium analyses were performed in Haploview v4.1 (http://www.broadinstitute.org/mpg/haploview/index.php). Insulin secretion was evaluated as either the mean (Utah cohort) or the area under curve for the 0–10 min acute insulin response to glucose (AIRG), glucose mediated glucose disposal (also called glucose effectiveness, SG) and insulin sensitivity (SI) was calculated from the FSIGT data using either the MinMod or MinMod Millennium programs [2,3,6,18]. Disposition index (DI), a measure of the ability of the β-cell to compensate for insulin sensitivity, was calculated as SI × AIRG (for units and definitions see foot note of Table 2). Genotypic effects on glucose homeostasis traits (SI, SG, AIRG and DI) were tested using mixed effect, general linear regression models (GLM) implemented in SPSS v.12 for Windows (SPSS Inc., Chicago, IL)., and age, body mass index (BMI), gender, genotype, protocol (tolbutamide or insulin), and diagnosis (IGT or glucose tolerant) were included as factors and covariates, as appropriate. To account for the relatedness of sibships from the Utah families, family number was included as a random factor in the GLM univariate analyses as described previously [1]. BGLAP expression between common homozygotes and C allele carriers groups was tested by Student’s T-test. All skewed variables including gene expression were ln-transformed to normality.
Table-2.
Association of marginal means for metabolic traits including FSIGT measures with genotype of BGLAP Polymorphisms rs34702397 (R94Q)
| rs34702397 | African American(Arkansas) | ||
| GG | GA | P | |
| N | 141 | 16 | |
| SI | 4.79 (4.12–5.57) |
3.45 (2.5–4.76) |
0.052 |
| AIRG | 2843 (2286–3536) |
2489 (1589–3967) |
0.58 |
| DI | 1354 (1071–1715) |
844 (509–1398) |
0.073 |
| SG | 0.017 (0.015–0.019) |
0.012 (0.009–0.016) |
0.031 |
| BMI | 29.3 (28.1–30.7) |
30.1 (27.3–33.2) |
0.62 |
Marginal means are shown after adjustment for age, gender and BMI; significance is based on a general linear model and p values are shown without correction for multiple testing. All means are transformed back to the linear scale from Ln-transformed values; 95% confidence intervals are provided in parenthesis. SI, insulin sensitivity index from MinMod in (pmol/l) min−1. AIRG in the Utah Caucasian study is from the mean 2–10 min post challenge insulin excursion (pmol/l); for Arkansas Caucasian and African American studies, AIRG is the area under the curve from 0 to 10 min, converted to pmol/l. DI, Disposition index (SI × AIRG) has no units. Values from the Utah sample are × 10−2. DI values for Arkansas samples are taken from the MinMod Millenium output. Based on the different calculation of AIRG the Utah and Arkansas DI values are not directly comparable. SG, glucose mediated glucose disposal (glucose effectiveness), is a measure of the ability of glucose to promote its own uptake (min−1). BMI, Body mass index (kg/m2). Data for the A92A polymorphism are not shown.
Results
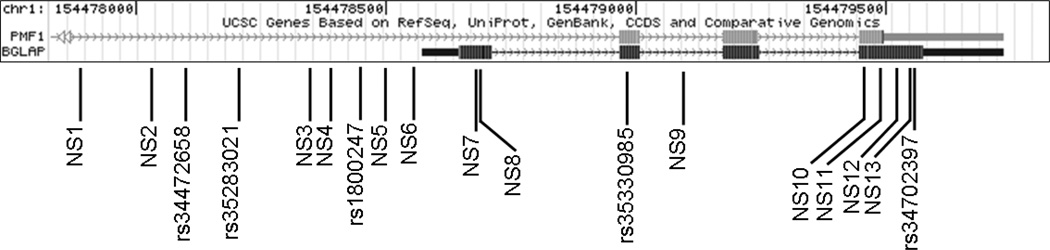
The BGLAP gene spans 1162 bp on chromosome 1q and includes four exons. We observed 18 SNPs (Figure 1), of which 13 were novel and observed only in African Americans (Supplementary Data, Table 2S). Only the putative promoter SNP rs1800247 was observed in both cohorts, whereas the nonsynonymous SNP rs34702397 (R94Q) was observed only in African Americans and synonymous SNP rs35330985 only in Caucasians. Because all SNPs other than rs1800247 were uncommon, we lacked the power to examine most variants. We tested rs1800247 in both populations, and coding SNPs rs34702397 (R94Q) and A92A (novel) in African American individuals. All SNPs were in Hardy Weinberg equilibrium, and no SNP was associated with T2D in either population (Table 1). Genotype data for the 1q dense map [20] was available for 352 Caucasian and 414 African American samples. SNPs within BGLAP were in very low levels of linkage disequilibrium with other typed chromosome 1q SNPs (r2< 0.1; Supplementary Data, Figure 1S).
Figure-1. Structure of Osteocalcin (BGLAP) gene and location of SNPs detected by sequencing.
NS, Novel SNP; Genomic location based on NCBI36.1 build.
Table-1.
Case-control association analysis of BGLAP SNPs genotyped in Caucasian and African Americans
| SNP name |
Variant (D/d) |
Population | Minor allele frequency | Genotype (DD/Dd/dd) | ||||
|---|---|---|---|---|---|---|---|---|
| Control | T2D | P-value | Control | T2D | P-value | |||
| rs1800247 | T/C | C | 0.223 | 0.208 | 0.52 | 186/102/17 | 287/156/18 | 0.55 |
| AA | 0.186 | 0.189 | 0.87 | 124/63/4 | 241/118/11 | 0.85 | ||
|
NS13 (A92A) |
C/G | AA | 0.034 | 0.046 | 0.43 | 177/13/0 | 337/32/1 | 0.64 |
|
rs34702397 (R94Q) |
G/A | AA | 0.042 | 0.038 | 0.62 | 174/16/0 | 344/24/2 | 0.44 |
D, major allele; d, minor allele; C, Caucasian; AA, African American; P-value, Fisher’s exact test 2-tailed P value for genotypic and allelic association. NS, Novel SNP
SNP rs1800247 was not associated with SI, fasting glucose, AIRG, DI or SG in either population (Supplementary Data, Table 3S). In contrast, rs34702397 (R94Q) was nominally associated with both SI (uncorrected p=0.052) and SG (uncorrected p=0.031) in African American individuals, but not with AIRG or DI (Table 2). No SNP was associated with obesity measures (BMI, waist: hip ratio, and percent body fat).
We examined the effects of minor allele carrier status for the putative promoter SNP rs1800247 on BGLAP expression in transformed lymphocytes. Transcript levels did not differ by genotype among cell lines from Caucasian individuals (Supplementary Data, Figure 2S).
Discussion
The identification of OCN resulted from the search for a bone-derived hormone that regulated energy metabolism in mice [12]. OCN is encoded by the 1162 bp, four exon BGLAP gene on human chromosome 1. OCN contains three glutamic acid residues in exon 4 (68, 72, and 75) that undergo carboxylation to γ-carboxyglutamic acid (Gla). Only uncarboxylated OCN induced expression of adiponectin in adipocytes and insulin and cyclinD1 in pancreatic islets [13]. In humans, γ-carboxylation is controlled by the vitamin K dependent enzyme γ-glutamyl carboxylase (GGCX). Functional polymorphisms of GGCX and the vitamin K epoxide reductase (VKORC1) gene were associated with uncarboyxlated OCN levels [17, 24]. Transcriptional regulation of BGLAP in humans is controlled by promoter binding of a complex that includes the transcription factor RUNX2, coactivator CBP/p300, and corepressor HDAC3 at the promoter [15, 21]. Several human studies also showed a significant negative correlation of serum OCN with fasting plasma glucose, fasting insulin, insulin resistance determined by the homeostasis model (HOMA), HbA1c, body mass index, percent and body fat, as well as a positive correlation with adiponectin [7, 9, 11, 19]. Thus, BGLAP variants might contribute to T2D susceptibility by altering expression, OCN levels, or γ-carboxylation. To our knowledge this is the first study to evaluate the role BGLAP variation with susceptibility to T2D or metabolic traits.
Despite exhaustive screening in two populations, we identified only one common SNP. Based on available published studies and linkage disequilibrium calculations from the present study, neither GWAS studies [22, 23] nor the chromosome 1q mapping [20] were likely to evaluate BGLAP adequately. Nonetheless, we find little variation in this region, and little evidence for an association with either T2D or related traits. We find nominal evidence for an association of the nonsynonymous SNP rs34702397 (R94Q) with SI and SG in African American individuals without correction for multiple testing. Clearly with Bonferroni correction, which is likely overly conservative given correlations between traits and between SNPs, these associations would no longer be significant. Furthermore, given the low frequency of this variant, confirmation is required in a much larger sample.
This study has some limitations. First, the majority of SNPs were rare. Hence, a much larger population would be required to detect a role in diabetes risk, particularly if the odds ratios are below 1.5 as might be expected from available data. Second, for effect sizes typical of T2D genes other than TCF7L2 [25], we had limited power. Hence, we cannot exclude a role for rs1800247 with an odds ratio of 1.2 or less, typical of many other T2D susceptibility genes. Third, we did not have a measure of OCN or uncarboxylated OCN in our populations. Association of SNPs with OCN levels might be helpful. We did not find evidence for an association with gene expression and no BGLAP SNP was predicted to alter γ-carboxylation. Although BGLAP is expressed in non-osseous tissues such as transformed lymphocytes, it lacks proper splicing [10] and thus may not reflect transcript levels in other tissues such as osteoblasts.
In summary, based on our analysis of a cohort with adequate size to detect a risk factor with an odds ratio over 1.5, BGLAP variants are unlikely to be a major risk factors for T2D and are unlikely to contribute to the 1q21 linkage signal in Caucasians. However, the arginine to glutamine change at residue 94 of exon 4 lies near the γ-carboxylation site may alter glucose homeostasis traits in African Americans and needs confirmation in a larger metabolic study.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health/NIDDK (DK039311). Metabolic studies partially supported by M01RR14288 from National Center for Research Resources (NIH) to the University of Arkansas for Medical Sciences (UAMS). We thank the UAMS Clinical Research Center nursing staff and laboratory for invaluable support in conducting these studies. We acknowledge Oksana Hackney for subject recruitment, Kurt Langberg for technical assistance and Dr. Ashis Mondal for characterization and cloning of BGLAP transcript from transformed lymphocytes.
References
- 1.Allison DB, Heo M, Kaplan N, Martin ER. Sibling-based tests of linkage and association for quantitative traits. Am. J. Hum. Genet. 1999;64:1754–1763. doi: 10.1086/302404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN. MINMOD Millennium: a computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther. 2003;5:1003–1015. doi: 10.1089/152091503322641060. [DOI] [PubMed] [Google Scholar]
- 3.Das SK, Sharma NK, Chu WS, Wang H, Elbein SC. Aryl hydrocarbon receptor nuclear translocator (ARNT) gene as a positional and functional candidate for type 2 diabetes and prediabetic intermediate traits: Mutation detection, case-control studies, and gene expression analysis. BMC Med Genet. 2008;9:16. doi: 10.1186/1471-2350-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das SK, Elbein SC. The search for type 2 diabetes susceptibility loci: the chromosome 1q story. Curr Diab Rep. 2007;7:154–164. doi: 10.1007/s11892-007-0025-3. [DOI] [PubMed] [Google Scholar]
- 5.Das SK, Elbein SC. The Genetic Basis of Type 2 Diabetes. Cellscience. 2006;2:100–131. doi: 10.1901/jaba.2006.2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbein SC, Hasstedt SJ, Wegner K, Kahn SE SE. Heritability of pancreatic beta-cell function among nondiabetic members of Caucasian familial type 2 diabetic kindreds. J Clin Endocrinol Metab. 1999;84:1398–1403. doi: 10.1210/jcem.84.4.5604. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Real JM, Izquierdo M, Ortega F, Gorostiaga E, Gómez-Ambrosi J, Moreno-Navarrete JM, Frühbeck G, Martínez C, Idoate F, Salvador J, Forga L, Ricart W, Ibañez J. The relationship of serum osteocalcin concentration to insulin secretion, sensitivity, and disposal with hypocaloric diet and resistance training. J Clin Endocrinol Metab. 2009;94:237–245. doi: 10.1210/jc.2008-0270. [DOI] [PubMed] [Google Scholar]
- 8.Ferron M, Hinoi E, Karsenty G, Ducy P. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proc Natl Acad Sci U S A. 2008;105:5266–5270. doi: 10.1073/pnas.0711119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Im JA, Yu BP, Jeon JY, Kim SH. Relationship between osteocalcin and glucose metabolism in postmenopausal women. Clin Chim Acta. 2008;396:66–69. doi: 10.1016/j.cca.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Jung C, Ou YC, Yeung F, Frierson HF, Jr, Kao C. Osteocalcin is incompletely spliced in non-osseous tissues. Gene. 2001;271:143–150. doi: 10.1016/s0378-1119(01)00513-3. [DOI] [PubMed] [Google Scholar]
- 11.Kanazawa I, Yamaguchi T, Yamamoto M, Yamauchi M, Kurioka S, Yano S, Sugimoto T. Serum osteocalcin level is associated with glucose metabolism and atherosclerosis parameters in type 2 diabetes mellitus. J Clin Endocrinol Metab. 2009;94:45–49. doi: 10.1210/jc.2008-1455. [DOI] [PubMed] [Google Scholar]
- 12.Lee NK, Karsenty G. Reciprocal regulation of bone and energy metabolism. Trends Endocrinol Metab. 2008;19:161–166. doi: 10.1016/j.tem.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyssenko V, Groop L. Genome-wide association study for type 2 diabetes: clinical applications. Curr Opin Lipidol. 2009;20:87–91. doi: 10.1097/MOL.0b013e32832923af. [DOI] [PubMed] [Google Scholar]
- 15.Makita N, Suzuki M, Asami S, Takahata R, Kohzaki D, Kobayashi S, Hakamazuka T, Hozumi N. Two of four alternatively spliced isoforms of RUNX2 control osteocalcin gene expression in human osteoblast cells. Gene. 2008;413:8–17. doi: 10.1016/j.gene.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nimptsch K, Nieters A, Hailer S, Wolfram G, Linseisen J. The association between dietary vitamin K intake and serum undercarboxylated osteocalcin is modulated by vitamin K epoxide reductase genotype. Br J Nutr. 2009;101:1812–1820. doi: 10.1017/S0007114508131750. [DOI] [PubMed] [Google Scholar]
- 18.Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986;23:113–122. doi: 10.1016/0169-2607(86)90106-9. [DOI] [PubMed] [Google Scholar]
- 19.Pittas AG, Harris SS, Eliades M, Stark P, Dawson-Hughes B. Association between serum osteocalcin and markers of metabolic phenotype. J Clin Endocrinol Metab. 2009;94:827–832. doi: 10.1210/jc.2008-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prokopenko I, Zeggini E, Hanson RL, Mitchell BD, Rayner NW, Akan P, Baier L, Das SK, Elliott KS, Fu M, Frayling TM, Groves CJ, Gwilliam R, Scott LJ, Voight BF, Hattersley AT, Hu C, Morris AD, Ng M, Palmer CN, Tello-Ruiz M, Vaxillaire M, Wang CR, Stein L, Chan J, Jia W, Froguel P, Elbein SC, Deloukas P, Bogardus C, Shuldiner AR, McCarthy MI for International Type 2 Diabetes 1q Consortium. Linkage disequilibrium mapping of the replicated type 2 diabetes linkage signal on chromosome 1q. Diabetes. 2009;58:1704–1709. doi: 10.2337/db09-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roca H, Franceschi RT. Analysis of transcription factor interactions in osteoblasts using competitive chromatin immunoprecipitation. Nucleic Acids Res. 2008;36:1723–1730. doi: 10.1093/nar/gkn022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 23.Scott LJ LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sogabe N, Tsugawa N, Maruyama R, Kamao M, Kinoshita H, Okano T, Hosoi T, Goseki-Sone M. Nutritional effects of gamma-glutamyl carboxylase gene polymorphism on the correlation between the vitamin K status and gamma-carboxylation of osteocalcin in young males. J Nutr Sci Vitaminol (Tokyo) 2007;53:419–425. doi: 10.3177/jnsv.53.419. [DOI] [PubMed] [Google Scholar]
- 25.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Boström KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jørgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjögren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Wellcome Trust Case Control Consortium. Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.