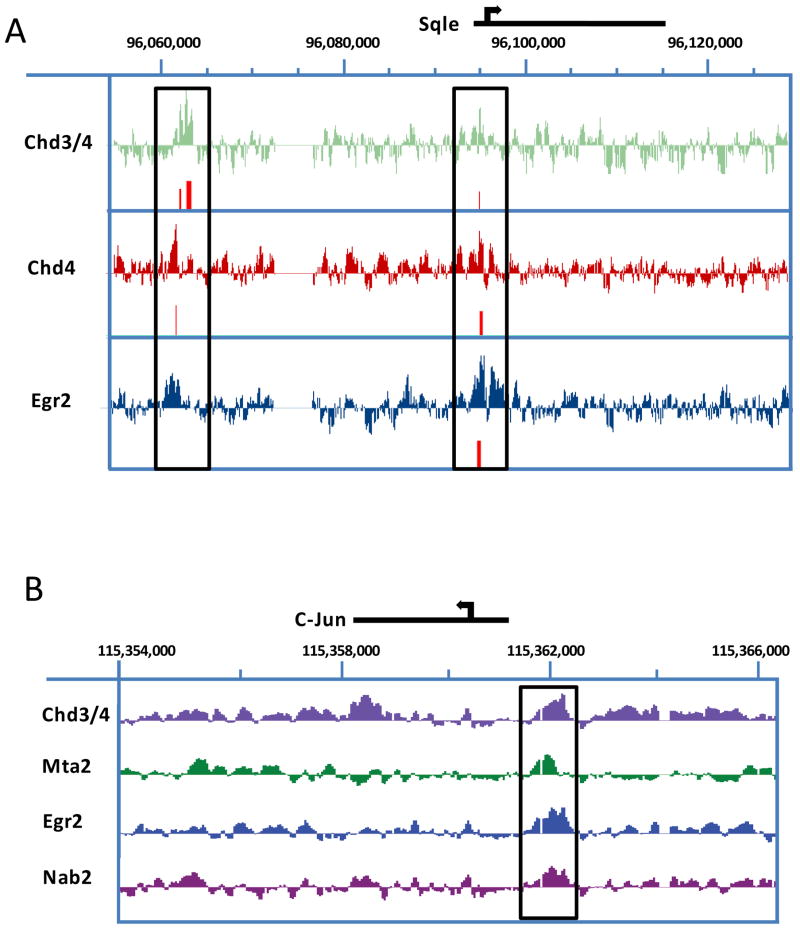
Figure 7. NuRD complex components and Egr2 assemble on both activated and repressed genes during myelination.
Chd4 binding is found on Sqle (A), and a negative regulator of myelination, c-Jun (B). ChIP-chip data for Egr2 (Jang et al., 2010) and two independent Chd4 antibodies were used to determine the binding location of Chd4 in P15 rat sciatic nerve. Data were analyzed using NimbleScan peak finding software. Peaks with a FDR< 0.05 are colored in red. ChIP-chip analysis for occupancy of Egr2, Chd4, and MTA2 on the Egr2-repressed gene, c-Jun (B), confirms binding of these proteins to the same loci upstream of the c-Jun promoter (box).