Figure 2.
Identification of a Rae1 binding motif in the PHR proteins.
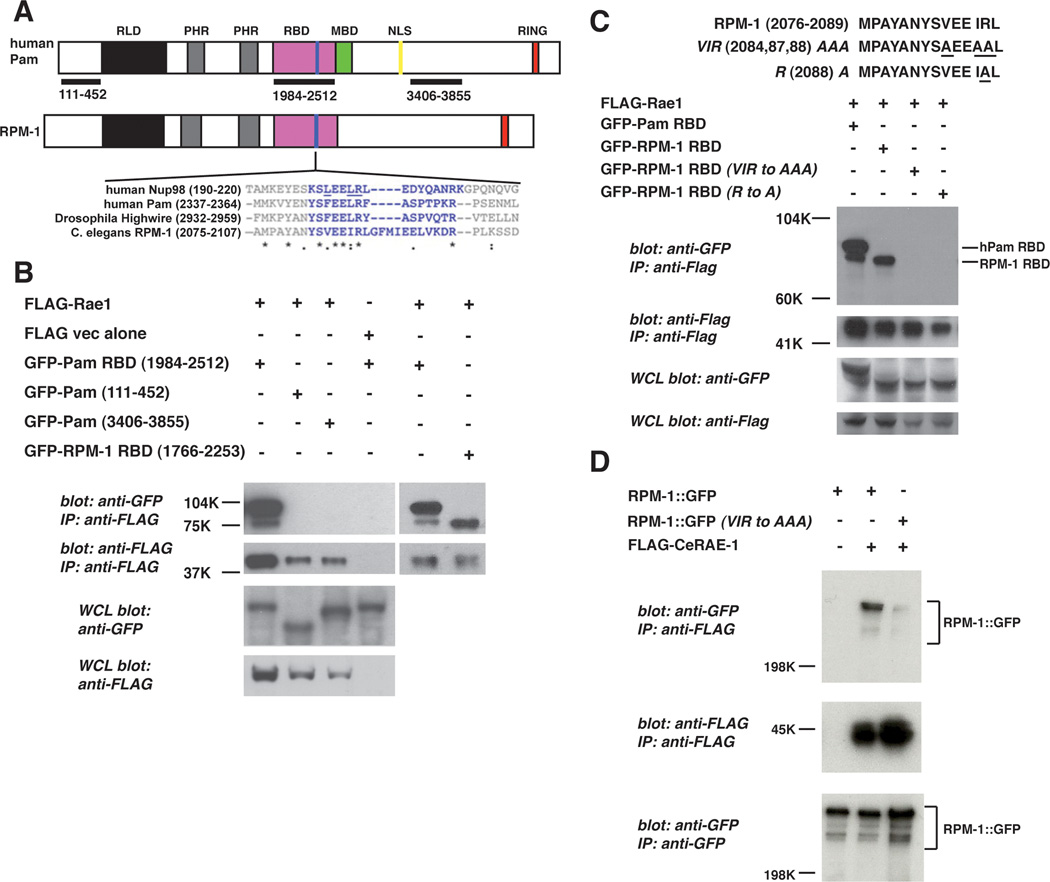
A, schematic representation of human Pam and C. elegans RPM-1. RLD (RCC-1 like domain), PHR (Pam/Highwire/RPM-1 family specific domains), RBD (Rae1 binding domain), MBD (myc binding domain), NLS (nuclear localization signal), and RING (RING-H2 ubiquitin ligase domain). Shown below is a sequence alignment of the known Rae1 binding protein, Nup98, and the PHR proteins. Residues highlighted in blue are conserved between Nup98 and other previously described Rae1 binding proteins. Residues in Nup98 that are underlined are essential for binding to Rae1. B, coIP of the Pam RBD or the RPM-1 RBD fused to GFP with Flag tagged rat Rae1. C, identification of point mutations in the RPM-1 RBD that reduce binding to FLAG-Rae1. D, coIP of RPM-1::GFP or RPM-1::GFP (VIR to AAA) point mutant with FLAG::RAE-1 from transgenic C. elegans protein lysates. Note that the RPM-1::GFP (VIR to AAA) point mutant has greatly reduced binding to FLAG::RAE-1 compared to wild-type RPM-1::GFP. A representative of at least three independent experiments is shown for B, C, and D. For D, at least two independently derived transgenic lines were analyzed for each genotype.