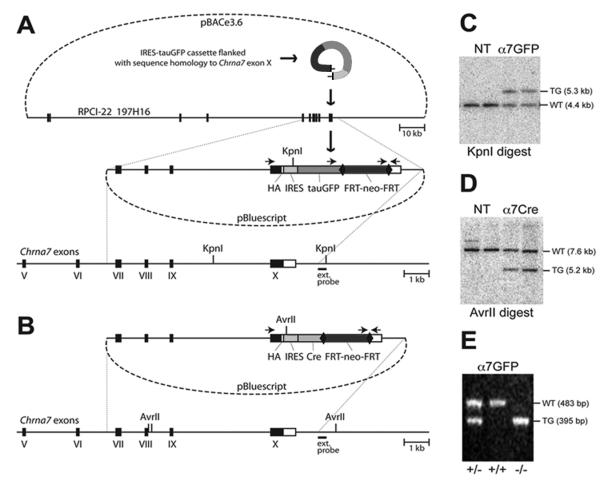
Figure 1.
Construction of the targeting vectors and generation of the corresponding mouse lines. (A) A diagram illustrating the production of Chrna7-IRES-tauGFP. In the first step, the IRES-tauGFP-FRT-neo-FRT cassette 1, including the HA epitope, was inserted in the 3′untranslated region (UTR) of Chrna7 (Methods). The external hybridization probe position, PCR priming sites and KpnI restriction sites are indicated. Numbered black boxes depict protein-coding exons. HA, hemagglutinin epitope; IRES, internal ribosomal entry site; FRT, recognition target sites of the FLP site-specific recombinase; neo, neomycin (kanamycin or G418 resistance); bovine tau polypeptide fusion to enhanced green fluorescent protein (tauGFP). (B) The Chrna7-IRES-Cre targeting vector was created by replacing tauGFP with the IRES-Cre-FRT-neo-FRT cassette 2. The external hybridization probe was as in (A). (C) Southern blot showing KpnI restriction fragment size up-shift from 4.4 kb (WT, wild type) to 5.3kb (TG, targeted allele) in the Chrna7-IRES-tauGFP targeted ES cell DNA. NT, negative ES clones harboring non-homologous recombination. (D) Chrna7-IRES-Cre and AvrII restriction analysis producing an anticipated downshift from 7.6 kb (WT) to 5.2 kb. (E) PCR genotyping of Chrna7-IRES-tauGFP mice in which the FRT-neo-FRT selection cassette was removed (Methods). The PCR data demonstrates the absence of wild type allele in normally segregating homozygous (-/-) Chrna7-IRES-tauGFP/IRES-tauGFP or Chrna7-IRES-Cre animals.