Abstract
Recombination occurs in many RNA viruses and can be of major evolutionary significance. However, rates of recombination vary dramatically among RNA viruses, which can range from clonal to highly recombinogenic. Here, we review the factors that might explain this variation in recombination frequency and show that there is little evidence that recombination is favoured by natural selection to create advantageous genotypes or purge deleterious mutations, as predicted if recombination functions as a form of sexual reproduction. Rather, recombination rates seemingly reflect larger-scale patterns of viral genome organization, such that recombination may be a mechanistic by-product of the evolutionary pressures acting on other aspects of virus biology.
Populations of RNA viruses habitually harbour abundant genetic variability, which is in large part due to a combination of high mutation rates and large population sizes1. Although RNA viruses were initially thought to experience only limited recombination, both experimental studies and analyses of the rapidly growing database of viral gene sequences have revealed not only that recombination occurs in many families of RNA viruses, but also that it can sometimes have a major impact on their evolution, emergence and epidemiology. Indeed, recombination has been associated with the expansion of viral host range2,3, increases in virulence4, the evasion of host immunity5 and the evolution of resistance to antivirals6. However, despite the growing evidence for the action of recombination in RNA viruses, the evolutionary reasons for its occurrence remain uncertain.
The process of recombination that takes place in RNA viruses corresponds to the formation of chimeric molecules from parental genomes of mixed origin. This process can occur either within a single genomic segment (in which case, it is often referred to as RNA recombination) or, for those viruses that possess segmented genomes, as exchange of entire genomic segments between viruses (FIG. 1). This exchange is usually termed reassortment. Although RNA recombination and reassortment are mechanistically very different, both require that two or more viruses infect the same host cell. In this Review, we describe the different mechanisms of recombination in RNA viruses and the evolutionary forces that shape the diversity of recombination rates.
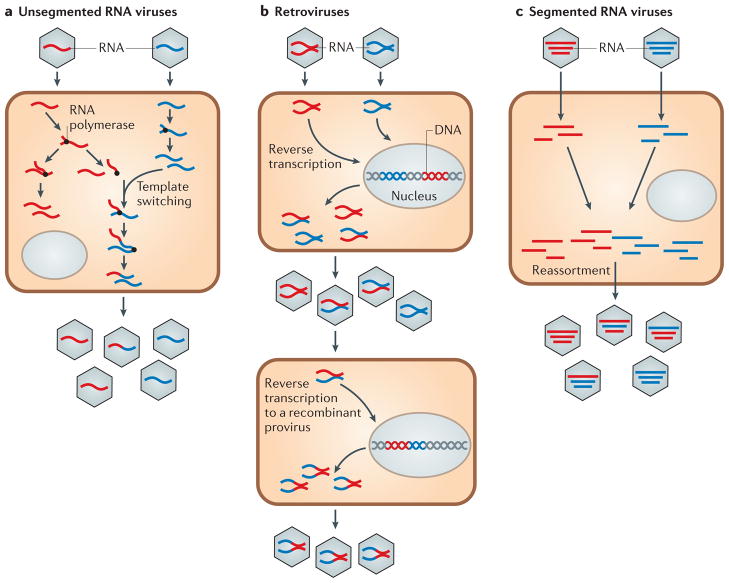
Figure 1. Generation of recombinant and reassortant RNA viruses.
a | Co-infection of a cell by genetically distinct viral strains can lead to the generation of recombinant viruses. This process can occur in both non-segmented viruses (as shown here) or within a segment of a segmented virus. b | Co-infection of a cell by genetically distinct strains of a retrovirus can lead to the generation of ‘heterozygous’ virus particles, after which a template-switching event can lead to a recombinant provirus. c | Co-infection of a cell by genetically distinct strains of a segmented virus can generate different combinations of reassortant progeny.
Models of recombination
In principle, RNA recombination can occur in all RNA viruses irrespective of whether their genomes are composed of single or multiple segments. Several models of RNA recombination have been proposed to explain the production of a genomic hybrid.
Copy choice recombination
The most widely accepted model of RNA recombination is ‘copy choice’ recombination7. In this process, the RNA polymerase that mediates viral replication — RNA-dependent RNA polymerase (RdRP) in most RNA viruses, and reverse transcriptase (RT) in retroviruses — switches from one RNA molecule (the donor template) to another (the acceptor template) during synthesis while remaining bound to the nascent nucleic acid chain, thereby generating an RNA molecule with mixed ancestry8–10 (FIG. 2). Various factors influence template switching, including the extent of local sequence identity between the RNA templates, the kinetics of transcription and secondary structure in the RNA11. Secondary structure may promote template switching through stalling of the RNA polymerase during replication and by facilitating the transfer of the polymerase and the nascent nucleic acid molecule onto the acceptor RNA12.
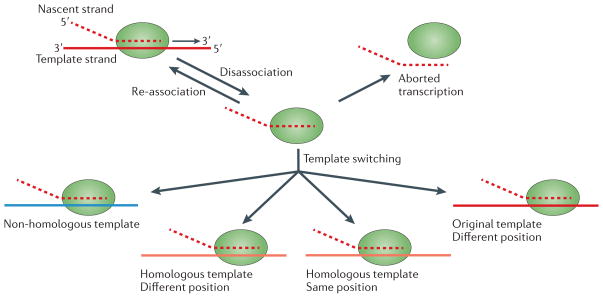
Figure 2. Potential consequences of a disassociation event during viral transcription.
Following disassociation of the viral polymerase and the nascent nucleic acid from the template, the polymerase has to find a template or the transcription process will abort. This re-association event can take place on the same template (red), at the same genomic position or at a different position. Alternatively, the polymerase can associate with a homologous template (orange), again either at the same genomic position or at a different position. Finally, re-association can take place on a non-homologous template, such as a cellular RNA (blue). Of all these possible RNA recombination events, those occurring at the same position on a homologous template are the most likely to generate functional progeny.
Template switching is thought to be guided by the sequence similarity between the nascent and the acceptor nucleic acid molecules13. Accordingly, RNA recombination is usually ‘homologous’, as it occurs most often between regions of high sequence similarity. Interestingly, the critical sequence similarity between the two parental sequences may be present close to, although not necessarily at, the crossover site. However, exchange between different, and hence genetically dissimilar, genomic regions or between non-related RNA molecules, leading to ‘non-homologous’ (or illegitimate) recombination, can also occur. As non-homologous RNA recombination involves regions with little sequence similarity, it will often produce deleterious genotypes, which is probably why it is observed far less frequently than homologous recombination (BOX 1).
Box 1. Are some viruses better adapted to deal with inevitable template switching?
If viral RNA polymerases harbour limited processivity and template switching may generate dysfunctional genomes29,116, the switching process might represent a liability for RNA viruses. Could specific features of some RNA viruses limit the potential risks that are associated with frequent template switching? Copy choice RNA recombination depends on sequence similarity, thus reducing the probability of illegitimate recombination and the chance of recombination among distantly related strains, both of which present high risks of generating deleterious genomes. However, sequence similarity is less relevant to copy choice recombination when the viral genome is covered by nucleoproteins, and this might explain why defective interfering particles are frequently found in negative-stranded RNA viruses. Similarly, although the process of dimerization in HIV does not favour the homodimerization of genomes, the compatibility of sequences at the dimerization initiation site is crucial to the formation of heterozygous particles and, hence, recombinants117. This may also explain, in part, the absence of recombination between HIV-1 and HIV-2, despite many cases of co-infection118, and a similar process could contribute to the nonrandom pattern reassortment that is observed in some segmented viruses119.
Finally, the risk of producing deleterious mutations by recombination is lowest when recombination breakpoints fall between rather than within genes, proteins or protein domains30. The observation that most intergenic and interprotein domains of HIV exhibit strong RNA structure is particularly striking in this respect, as these structures favour template switching in genomic regions where recombination is less likely to be disruptive120. Similarly, the conserved motifs that contribute to the specific template-switching events for discontinuous transcription in coronaviruses are also likely to enhance template switching in these intergenic regions, in turn limiting the chances of intraprotein recombination (in a phenomenon that is reminiscent of reassortment).
Direct experimental approaches and bioinformatic methods that search for incongruence in phylogenetic trees have identified instances of both homologous and non-homologous RNA recombination in various RNA viruses14. Furthermore, defective interfering particles, which are virions that contain truncated viral genomes, have been observed in most viral families15,16. These truncated viral genomes are a consequence of the low processivity of RNA polymerases and are probably produced through a copy choice mechanism17.
Non-replicative RNA recombination has also been demonstrated experimentally18–20, albeit at a much lower frequency than copy choice recombination. During this process, recombining RNAs are cleaved at specific points and ligated to form hybrid molecules. Several enzyme-catalysed mechanisms have been proposed to explain non-replicative RNA recombination, and RNA secondary structure rather than sequence similarity is thought to be the major factor mediating this process.
Reassortment
Reassortment is the second form of genetic exchange that has been described in RNA viruses. It is restricted to viruses that possess segmented genomes, and involves packaging of segments with different ancestry into a single virion. As with RNA recombination, reassortment requires that a cell be infected with more than one virus (FIG. 1). Although reassortment does not require the physical proximity of parental genomes during replication, the packaging process that results in reassortant viruses may not be entirely random21–24.
It is possible that segmented viruses arose following the co-infection of a single cell by two or more viruses, which then evolved to function together through complementation. For example, segmented plant viruses frequently co-infect a common host25 and have a high multiplicity of infection (MOI)26,27, often leading to mixed infections. Indeed, many positive-sense single-stranded RNA ((+)ssRNA) viruses that target plants have segmented genomes which are encapsidated into separate particles; these so-called multicomponent viruses include those in the families Bromoviridae and Comoviridae, and some members of the families Virgaviridae and Secoviridae. Alternatively, segmented genomes may have evolved directly from unsegmented ones. In particular, defective interfering particles are commonly produced during the in vitro passage of RNA viruses at a high MOI, and some of these defective interfering particles could remain infectious through complementation28 and could conceivably represent incipient genome segments.
RNA recombination and reassortment may also differ in their likelihood of generating deleterious genotypes. Both processes may bring together incompatible mutations in a single genome29. This risk increases with phylogenetic distance between the parental strains but also varies according to the specific position of the recombination site with respect to genes, proteins and protein domains30. Hybrid proteins are more likely to fold and be functional when the recombination breakpoint occurs at a position that minimizes the number of disrupted interactions. Although reassortment by-passes this risk, as it involves the replacement of entire transcriptional units (that is, segments), the possibility of disrupting co-evolved intermolecular interactions remains31,32 and might be especially strong in the case of segments that encode components of multiprotein complexes.
Recombination frequencies in RNA viruses
RNA recombination and reassortment occur at highly variable frequencies in RNA viruses, although there are few instances in which precise rates of recombination per nucleotide or genome have been determined (BOX 2). For example, recombination appears to occur frequently in some retroviruses33 — most notably HIV, which has an estimated recombination rate of between 1.38 × 10−4 and 1.4 × 10−5 per site per generation34,35 — and in some (+)ssRNA viruses, such as enteroviruses (of the family Picornaviridae)36 and viruses of the families Coronaviridae37, Bromoviridae38 and Potyviridae39,40. However, recombination appears to be far less frequent in other families of (+)ssRNA viruses, including the Flaviviridae, in which only occasional instances have been reported41–44. For example, in the case of hepatitis C virus (HCV), recombination frequencies of only 4 × 10−8 per site per generation were reported during co-infection experiments45. In addition, recombination has thus far not been detected in a number of (+)ssRNA viruses, including members of the families Barnaviridae, Leviviridae, and Narnaviridae and the genera Benyvirus, Cilevirus and Idaeovirus, although in some cases this may reflect the limited amount of data available.
Box 2. Measuring recombination rates in RNA viruses.
Recombination rates in RNA viruses have been measured in two different ways: the intrinsic rate of template switching that occurs during replication, and the recombination rate that can be inferred at the population level. Unfortunately, large-scale comparative studies of recombination rates using either approach have yet to be undertaken. To accurately measure the frequency of template switching, recombinant products need to be accessed soon after their generation, before any form of selection can take place. Although they were initially measured in vitro, recombination rates are now obtained from single-cycle experiments in co-infected cells in culture. These experiments provide valuable insights into the frequency and mechanisms of recombination, but they do not necessarily mimic natural systems, in which the frequency of co-infection is unknown. The frequency of co-infection depends on many factors, including the size of the viral population, the frequency of different variants in the population and the likelihood of mixed infection. Recombination rates may also be estimated in live hosts112, as has been carried out in natural HIV infections34.
Methods that estimate the recombination rate at the population level are based on gene sequence analysis8 and necessarily exclude deleterious recombinant forms that have been removed by purifying selection. Some of these approaches explore linkage disequilibrium in population-wide genetic data sets113,114, whereas others rely on coalescent approaches to provide estimators of the population recombination rate115. The power of these approaches is that they allow a standardized framework for comparing recombination rates. However, they are limited in that the signal of recombination is sometimes difficult to distinguish from particular mutation patterns or from other factors such as geographical structure in the data, and that they rely heavily on the sample of sequences used in the analysis, which may be both small and biased.
Current data indicate that recombination is even less frequent in (−)ssRNA viruses46,47, although (−)ssRNA viruses with segmented genomes can still undergo reassortment. For example, analyses of thousands of gene sequences from influenza A virus isolates have revealed only sporadic evidence for homologous recombination in this virus, and all these instances are open to alternative explanations48. However, reassortment clearly occurs very frequently in this virus49–51. As discussed below, the low frequency of RNA recombination in (−)ssRNA viruses seems to reflect specific aspects of their life cycle and therefore provides important clues as to the forces controlling the evolution of recombination in RNA viruses as a whole.
Importantly, the extensive variation in recombination frequency among RNA viruses seems to correspond to a number of fundamental biological differences between them. For example, viruses that generate persistent rather than acute infections, such as HIV, may have higher rates of recombination because a single host has an increased chance of acquiring mixed infections. Similarly, ecological processes, such as whether hosts meet sufficiently regularly for co-infection to occur, must also influence recombination rates to some extent. Certain genome structures also clearly facilitate recombination. For example, the genomic organization of retroviruses favours the occurrence of genetic exchange. Retrovirus virions carry two RNA molecules, making them ‘pseudodiploid’. As a consequence, viruses with different ancestries that simultaneously infect a host cell may be packaged together, producing ‘heterozygous’ virions and thereby enabling the production of genetically distinct progeny through copy choice recombination. It has been estimated that the RT of HIV-1 switches template 2–20 times per replication cycle, thereby exceeding the rate of mutation measured per nucleotide in this virus52,53, and making it one of the most recombinogenic of all RNA viruses. However, not all retroviruses recombine with the frequency of HIV. For example, in murine leukemia virus (MLV) and gammaretroviruses in general, recombination is 10–100 times less frequent than in HIV, despite having very similar copy choice rates.
Similarly, there is extensive variation in the rate of reassortment among viruses. Specifically, whereas many segmented viruses (such as hantaviruses54, Lassa virus55 and tenuiviruses56) exhibit relatively low levels of reassortment, this process appears to occur much more frequently in other viruses. For example, in the case of influenza A virus, at least 2–3 reassortment events occur per year57, and 2.7–5.4% of rotaviruses were found to be reassortant58,59. Indeed, in both influenza A virus and rotavirus A, the reassortment of different gene segments encoding viral envelope or surface proteins — specifically, the haemagglutinin (HA) and neuraminidase (NA) of influenza A viruses, and VP7 and VP4 proteins of rotavirus A (which define the G and P genotypes, respectively) — is associated with the evasion of host immunity and, sometimes, the occurrence of epidemics58,60,61. Even higher rates of reassortment may characterize the cystoviruses, as the extent of linkage disequilibrium (LD) between viral segments is close to zero, as expected for a completely sexual population62.
Recombination as a form of sexual reproduction
Although the wide range of recombination rates is one of the most interesting issues in the study of viral evolution, it is also one that has received little attention. As we discuss below, the central question is whether recombination functions as a form of sex in RNA viruses, so that it is selectively favoured as a means to create or purge genetic variation, or whether it is a secondary by-product of the replication process, such that the variation in its rate reflects the diversity of the genome structures and life histories exhibited by RNA viruses. Most of the discussion about the factors that might explain the evolution of recombination in RNA viruses has focused on the two major advantages of recombination over asexual evolution: recombination accelerates the rate at which advantageous genetic combinations are produced and allows a more efficient removal of deleterious mutations (FIG. 3).
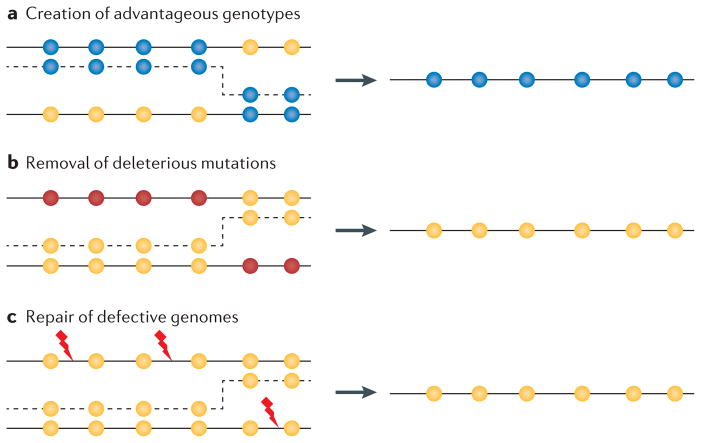
Figure 3. Evolutionary consequences of recombination.
Depending on the acceptor and donor genotypes, and the position of the template switch, recombination can have several positive effects on the genome. Yellow circles indicate wild-type loci. a | Recombination can create advantageous combinations of mutations (blue circles) that increase the rate of adaptive evolution compared with mutation alone, or it can disassociate advantageous and deleterious mutations, allowing the former to spread. b | Recombination can remove deleterious mutations (red circles) and restore the wild-type (fit) genotype, which can lead to a selective advantage for recombination if deleterious mutations occur frequently enough and interact synergistically. c | Recombination can also generate a functional genome from damaged parental molecules. Genetic damage, such as strand breaks or oxidative base modifications, are represented by red lightning symbols.
Rapid creation of advantageous genotypes
Although recombination undoubtedly accelerates the rate at which advantageous genetic combinations are produced and frees advantageous mutations from deleterious genomic baggage63, it is unclear whether this provides sufficient selective advantage to make it the sole explanation for the evolution and maintenance of sexual reproduction64. It is also unlikely that an elevated rate of adaptive evolution is sufficiently beneficial to populations of RNA viruses for this to be the main reason for the existence of recombination. For example, although recombination can clearly assist the development of drug resistance in HIV6,65–68, most cases of antiviral resistance in this virus have been associated with the accumulation of single point mutations. Similarly, although both drug resistance and antigenic escape occur commonly in HCV, recombination has been observed only sporadically in this virus69–71 and is thus unlikely to be of selective importance. It may be that the mutation rates are normally so high in RNA viruses (at between 10−6 and 10−4 mutations per nucleotide per cell infection72) and the population sizes often so large (both within hosts and at the epidemiological scale) that advantageous combinations of mutations are regularly generated without the assistance of recombination.
Another aspect of the effect of recombination on the rate of adaptive evolution is ‘clonal interference’. This refers to that fact that advantageous mutations will compete — and hence interfere — with each other in asexual populations such that only one beneficial mutation can be fixed at any one time73. By contrast, because advantageous mutations in sexual populations can arise in different individuals and then recombine into a single genome, adaptive evolution can proceed at a higher rate. Although clonal interference has been experimentally demonstrated in some asexual RNA viruses at large population sizes73–75, seemingly clonal (−)ssRNA viruses are commonplace and can cause epidemics in many species; clonal interference is therefore unlikely to be a major selection pressure for the evolution of recombination.
Purging deleterious mutations
According to a second theory, recombination constitutes an efficient way of removing deleterious mutations. The simplest form of this theory, described by Muller’s ratchet, states that the continual and irreversible accumulation of deleterious mutations in small asexual populations leads to a progressive decrease in fitness, and that genetic drift accelerates the loss of mutation-free individuals76. Sexual populations would be buffered from this fitness loss. As Muller’s ratchet has been demonstrated to occur in experimental populations of RNA viruses75,77–81, it may describe several aspects of viral evolution. However, other than at the population bottlenecks that probably occur at inter-host transmission, it is debatable whether the population sizes of RNA viruses are small enough for Muller’s ratchet to be a regular occurrence.
A more popular theory for the origin of sexual reproduction based on the purging of deleterious mutations, known as the mutational deterministic hypothesis, considers infinite population sizes82,83. The mutational deterministic hypothesis requires the rate of deleterious mutations per genome replication (U) to be greater than 1. It also requires these deleterious mutations to be subject to synergistic epistasis, so that their combined effect on fitness is greater than that expected from their individual effects83. Several studies have found that the rate of acquisition of deleterious mutations is indeed high in RNA viruses, such that U > 1 may be a realistic occurrence84–86. However, although deleterious mutations occur frequently, there is no good evidence for the occurrence of frequent synergistic epistasis in RNA viruses, as most of the described epistatic interactions are antagonistic87–90. A high frequency of antagonistic epistasis is expected in RNA viruses because of the lack of genetic redundancy in their highly constrained genomes, such that individual proteins often perform multiple functions. Additional evidence against the mutational deterministic theory is that the burden of deleterious mutations is seemingly high in all RNA viruses, indicating that it does not depend on genome structure or, hence, the propensity for recombination86.
In summary, it seems unlikely that recombination has evolved as a means by which RNA viruses can purge deleterious mutations. Rather, the population sizes of RNA viruses may be so large that sufficient viable progeny are produced every generation to guarantee survival. In addition, large population sizes mean that the accumulation of deleterious mutations can be offset by frequent back and compensatory mutations. RNA viruses may therefore possess a population-scale robustness that protects them from the accumulation of deleterious mutations91.
Finally, it is important to note that there are also evolutionary costs associated with recombination in RNA viruses, as it is likely to increase both the degree of competition within a host92 and the extent of complementation. Complementation is particularly important because it enables defective viruses to parasitize the fully functional viruses that co-infect the same host cell, thereby allowing deleterious mutations to remain in viral populations for longer time periods. Co-infection is a prerequisite for both recombination and complementation. Interestingly, protection from co-infection has been described for numerous plant viruses93, and mechanisms that limit the co-infection of individual cells by multiple viruses have been documented in a number of other viruses, including retroviruses94, pestiviruses95 and alphaviruses96,97, some of which recombine frequently.
Non-sexual evolution of viral recombination
In contrast to the theories described above, various other theories of the evolution of recombination in RNA viruses do not consider it a form of sexual reproduction.
Repair of genetic material
One theory states that the adaptive value of recombination comes from its ability to repair genetic damage98. Indeed, early work on recombination in retroviruses suggested the existence of a ‘forced copy choice’ model of recombination, in which the template switch occurs when a break in the RNA template forces the RT to seek an alternative and functional template99. However, replication occurs equally often on unbroken and on broken templates100, and experimentally induced genetic damage has not been clearly associated with higher recombination rates101. Moreover, if genetic damage is the driving force behind recombination, the common exposure of viruses to oxidative stress is at odds with the strong disparities in their rates of recombination. In theory, recombination as a form of repair could be a potent mechanism in viruses with diploid or pseudo-diploid genomes, such as HIV. However, the process clearly relies on high rates of multiple infection, and as this is a feature that cannot be guaranteed for all viruses, it is unlikely that there would be sufficient pressure for recombination to be selected as a repair mechanism.
Recombination as a by-product of genome organization
Several theories for the evolution of recombination in RNA viruses argue that rather than being selected for its direct fitness benefits, as is proposed by all theories in which recombination functions as a form of sex, recombination is in fact a by-product of the processivity of RNA polymerases. Under this hypothesis, the observed variation in the recombination rates is largely determined by differences in the genome organization and life cycles of RNA viruses (TABLE 1).
Table 1.
Differing genome organizations and replication strategies of RNA viruses
| Virus family | Genome | Segmentation | Genome replication strategy | Protein expression strategy |
|---|---|---|---|---|
| Bromoviridae | Positive sense | Three segments | Makes negative-sense copies of segments | Makes genomic and subgenomic RNA (from an internal promoter) |
| Bunyaviridae | Negative sense | Three segments | Makes positive-sense copies of segments (the polymerase templates are RNPs) | Uses single-segment ORFs and then cleaves the resultant polyproteins |
| Coronaviridae | Positive sense | Unsegmented | Makes a negative-sense copy of the genome | Produces mRNA from genomic and subgenomic negative-sense RNA through discontinuous transcription |
| Flaviviridae | Positive sense | Unsegmented | Makes a negative-sense copy of the genome | Uses a single genome ORF and then cleaves the resultant single polyprotein |
| Retroviridae | Positive sense | Unsegmented (two copies) | Converts the genome to DNA, which integrates into the host genome | Uses the cellular machinery |
| Rhabdoviridae | Negative sense | Unsegmented | Makes a positive-sense copy of the genome (the polymerase templates are RNPs) | Processively transcribes each gene (the resultant mRNAs are not RNPs) |
| Togaviridae | Positive sense | Unsegmented | Makes a negative-sense copy of the genome | Makes genomic and subgenomic RNA (from an internal promoter) |
The basis of this theory is the notion that many aspects of genome organization in RNA viruses aim to control gene expression. Specifically, a major challenge for all RNA viruses is to control the levels of each protein that they produce. Many unsegmented (+)ssRNA viruses control gene expression initially at the level of translation, as this is the first step in the viral life cycle. Translation often results in a single large polyprotein that then needs to be proteolytically cleaved into individual proteins. Although this replication strategy is efficient and allows the naked RNA that is extracted from virions to be infectious in the absence of a co-packaged viral polymerase, it also means that equal amounts of each protein are produced; any difference in protein abundance must be achieved through differential polyprotein cleavage. This constraint can be overcome by dividing the viral genome into separate ‘transcriptional units’ that offer greater control over gene expression1. The (+)ssRNA viruses have achieved this in a variety of ways, including the use of subgenomic RNAs (as seen in the families Tymoviridae and Togaviridae and in the genus Sobemovirus), the use of ribosomal frame-shifting (as employed by the order Nidovirales) and the evolution of distinct genomic segments. In the case of segmented viruses, reassortment would then occur through the normal packaging mechanism and, hence, as a by-product of co-infection by two segmented viruses, although this does not preclude the production of selectively beneficial genetic configurations. The observation that genome segmentation is more common in (+)ssRNA viruses than in (−)ssRNA viruses supports the theory that the necessity of controlling gene expression is a key factor determining the evolution of genome organization, as (−)ssRNA viruses have more options than (+)ssRNA viruses for the control of gene expression at the level of transcription (see below). Similarly, the existence of polycistronic mRNAs in bacteria, in contrast to the monocistronic mRNAs of eukaryotes, may explain why fewer segmented viruses of bacteria have been described to date, although this may change with an increased sampling of the virosphere.
Improved control of gene expression may also explain the origin of (−)ssRNA viruses and their very low rates of recombination. In some respects, the existence of these viruses is puzzling, as they need to go through an additional transcription step before they can translate their proteins, and they also need a viral transcriptase (RdRP) to enter the host cell in addition to the RNA. However, such a life cycle opens up a powerful means to control gene expression at the level of transcription; transcription produces multiple mRNAs, which can form individual transcriptional units. Furthermore, the negative genome orientation removes the problem of having to use the same template for both translation and replication. It is also striking that the genome organizations of unsegmented (−)ssRNA viruses exhibit a common pattern, with the gene encoding the nucleocapsid (N) protein being the first to be transcribed and the gene encoding the RdRP (the L protein) being the last to be transcribed. This gene order results in a strong gradient of transcription such that more N protein is produced than L protein, probably because the enzymatic function of the L protein requires fewer copies of the protein than the number required for the structural nucleocapsid. Such a conserved gene order, and one that correlates with the amount of protein product required, strongly suggests that natural selection is operating at the level of gene expression. Indeed, experimentally changing the gene order of (−)ssRNA viruses can result in major fitness reductions102.
The low levels of recombination in (−)ssRNA viruses appear to be a direct consequence of this form of genome organization. Specifically, the genomic and antigenomic RNA molecules in viruses of this type are quickly bound to multiple nucleoprotein subunits, as well as to other proteins, to form ribonucleoprotein (RNP) complexes from which viral replication and transcription can proceed. However, this tight complex of RNA and proteins lowers the probability of hybridization between complementary sequences in the nascent and acceptor nucleic acid molecules, and it is this hybridization that is required for the template switching that occurs during copy choice recombination. Furthermore, the specific recognition of RNP-bound RNA by the RdRP reduces the potential number of substrates for template switching. It is therefore not surprising that most of the defective interfering particles of (−)ssRNA viruses correspond to intramolecular recombination events that do not require a switch to another RNP template17. Intriguingly, ‘illegitimate’ recombination with a cellular mRNA has also been described in influenza A virus4. Together, these observations suggest that homologous recombination is possible in (−)ssRNA viruses but is hampered by a lack of suitable substrates. Indeed, although recombination in (−)ssRNA viruses is infrequent46, phylogenetic analyses have revealed a number of interesting cases in human respiratory syncytial virus103, Arenavirus104,105 and Ebolavirus106.
At the other end of the spectrum, recombination has been shown to be very frequent in (+)ssRNA corona-viruses such as murine hepatitis virus (MHV), for which up to 25% of the progeny of co-infected cells were found to be recombinant. Interestingly, the strategy of gene expression that is used by coronaviruses — discontinuous transcription — relies entirely on the template-switching property of the viral RdRP. Discontinuous transcription of the large unsegmented genomes of MHV and similar coronaviruses leads to the production of subgenomic negative-sense RNAs through a copy choice mechanism; these RNAs serve as templates for the production of mRNA107. This suggests that the RdRP of MHV is selected to efficiently mediate template-switching events and that the very high rates of recombination observed are a direct consequence of this particular strategy for controlling gene expression.
Last, in some retroviruses, most notably HIV, recombination rates are extremely high. The pseudodiploidy of these viruses facilitates recombination because two RNA molecules must be packaged in the same virion, thus increasing the likelihood of template switching owing to the physical proximity of the RNAs during replication. Furthermore, template switching is also an intrinsic component of the replication strategy of retroviruses. To integrate into the host genome, retroviruses convert their (+)ssRNA genome into a double-stranded (ds) DNA molecule. This generation of the dsDNA genome is not a straightforward conversion of positive-sense RNA into negative-sense DNA, followed by the synthesis of a positive-sense DNA complement; instead, two template switches, known as strong-stop strand transfers, are required to join and duplicate the long terminal repeats at the boundaries of the provirus. However, whereas strong-stop strand transfers occur only at specific positions, copy choice template switching may occur at any position in the retroviral genome.
The occurrence of pseudodiploidy and frequent template switching might suggest that these processes have been selected for to increase recombination rates in retroviruses, but recombination rates in fact differ greatly between retroviruses in a manner that reflects other aspects of viral biology. For example, genome dimerization for HIV probably occurs randomly in the cytoplasm. By contrast, genome dimerization for MLV takes place in the nucleus, close to the transcription sites, and leads mostly to self-associations rather than the associations between genetically different parental molecules that would be needed for recombination108. More generally, both diploidy and complementation allow deleterious mutations to be masked (in the case of diploidy, as recessive mutations), and this may provide an explanation for the evolution of diploidy109–111. Importantly, however, there is no evidence that the rate of deleterious mutation in HIV differs from that seen in ‘haploid’ (+)ssRNA and (−)ssRNA viruses.
Conclusions
Understanding the evolution of recombination remains one of the most challenging problems in biology. We suggest that it is optimistic to believe that a single explanation applies to all organisms and that the precise mechanisms of recombination must be understood in each case. In particular, although it is clear that recombination is a key aspect of sexual reproduction in most cellular species, such that its evolution can be discussed in terms of the generation and removal of specific types of mutation, we argue that this does not seem to be the case in RNA viruses. Indeed, it is striking that high levels of recombination appear to be a sporadic occurrence in RNA viruses, such that they cannot be universally advantageous, whereas theories for the evolution of sexual reproduction in eukaryotes attempt to explain recombination and clonality on the assumption that these are common and sporadic, respectively63. Rather, a review of the available data suggests that the differing rates of recombination and reassortment that characterize RNA viruses may reflect the mechanistic constraints that are associated with particular genome structures and viral life cycles. If this hypothesis is upheld, then recombination should be considered a mechanistic by-product of RNA polymerase processivity (a trait that varies according to the genomic architecture of the virus in question) and not as a trait that is optimized by natural selection for its own selective value, although it may on occasion generate beneficial genotypes (BOX 3). It is important to note that RNA viruses produce large numbers of progeny and that this, rather than recombination, is more likely to be the key to their evolutionary survival, as it buffers them from the adverse effects of the accumulation of deleterious mutations and regularly produces advantageous mutations. However, it is equally clear that our knowledge of recombination and its determinants remains patchy for most RNA viruses, such that far more data are needed for a definitive understanding of the evolution of recombination. For example, it will be important to accurately measure recombination rates in viruses that differ markedly in their strategies for controlling gene expression. Fortunately, the development of next-generation sequencing methods is likely to facilitate the acquisition of data that will lead to important new insights into the causes and consequences of recombination in this major class of infectious agent.
Box 3. Recombination and viral emergence.
For most RNA viruses, cross-species transmission is the most common way for a virus to enter a new host. Recombination could assist in this process because it enables viruses to explore a greater proportion of the sequence space than is accessible by mutation at any one time, thereby increasing the likelihood of finding a genetic configuration that facilitates host adaptation. Notably, many recently emerged human diseases are caused by RNA viruses that display active recombination or reassortment. Recombination and reassortment are also powerful ways for emerging viruses to acquire new antigenic combinations that may assist the process of cross-species transmission. The continual shuffling of genes that encode the haemagglutinin and neuraminidase envelope proteins of influenza A virus (a virus that is commonly associated with the occurrence of human pandemics) represents a powerful example of the benefits of recombination and reassortment for the virus50,60.
However, in most cases the emergence of a specific virus cannot be directly attributed to its ability to recombine. For example, although HIV-1 recombines at a high rate, there is no evidence that recombination assisted the cross-species transfer of the virus from the chimpanzee reservoir population into humans. In fact, there are few cases in which recombination seems to have directly resulted in viral emergence. One of the few examples is the alphavirus Western equine encephalitis virus, which was generated through recombination between a Sindbis-like virus and an Eastern equine encephalitis virus-like virus121. Similarly, a new coronavirus that emerged in turkeys is a recombinant infectious bronchitis virus that acquired a spike protein-encoding gene from another coronavirus122. Finally, the retrovirus Rous sarcoma virus is likely to have acquired pathogenicity through the recombination-mediated acquisition of a cellular oncogene123. In summary, the available data suggest that although recombination is sometimes directly helpful to the process of cross-species transmission, it is not a necessary precursor to successful viral emergence.
Acknowledgments
E.C.H. is supported in part by US National Institutes of Health grant R01GM080533.
Glossary
- Genomic segment
An independently replicating RNA molecule. RNA viruses can possess either a single segment (such that they are unsegmented) or multiple segments. Those with multiple segments may experience reassortment
- Defective interfering particles
Defective viruses (usually possessing long genome deletions) that compete, and hence interfere, with fully functional viruses for cellular resources
- Virions
The final mature virus particles containing the RNA genome and the full set of proteins
- Processivity
A measure of the average number of nucleotides added by a polymerase enzyme per association–disassociation with the template during replication
- Packaging
The process by which the nucleic acid genome and other essential virion components are inserted in the structural core or shell of a virus particle
- Complementation
The process by which a defective virus can parasitize a fully functional virus that is infecting the same cell; the defective virus ‘steals’ the proteins of the functional virus to restore its own fitness
- Multiplicity of infection
(MOI). The ratio of viruses to the number of cells that are infected
- Breakpoint
The site in the genome sequence at which a recombination event has occurred. Phylogenetic trees are incongruent on either side of the breakpoint
- Linkage disequilibrium
(LD). The nonrandom association between alleles at two or more loci, being indicative of a lack of recombination. Recombination reduces LD
- Clonal interference
The process by which beneficial mutations compete, and hence interfere, with each other as they proceed toward fixation
- Epistasis
An interaction between mutations such that their combined effect on fitness is different to that expected from their stand-alone effects. Depending on the nature of the deviation, epistasis can be either antagonistic (positive) or synergistic (negative)
- Genetic redundancy
The situation in which a specific phenotype is determined by more than one gene, such as members of multigene families
- Robustness
The constancy of a phenotype in the face of pressure from a deleterious mutation
- Provirus
The DNA form of a retroviral genome that is integrated into the genetic material of a host cell
- Genome dimerization
A non-covalent process by which retroviruses carry two RNA genomes in the virion
- Sequence space
All possible mutational combinations that are present in DNA or amino acid sequence data
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Edward C. Holmes’s homepage: http://www.cidd.psu.edu/people/ech15
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Holmes EC. The Evolution and Emergence of RNA Viruses. Oxford Univ. Press; New York: 2009. [Google Scholar]
- 2.Brown DW. Threat to humans from virus infections of non-human primates. Rev Med Virol. 1997;7:239–246. doi: 10.1002/(sici)1099-1654(199712)7:4<239::aid-rmv210>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs MJ, Weiller GF. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci USA. 1999;96:8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khatchikian D, Orlich M, Rott R. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature. 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 5.Malim MH, Emerman M. HIV-1 sequence variation: drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 6.Nora T, et al. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81:7620–7628. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai MM. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aaziz R, Tepfer M. Recombination in RNA viruses and in virus-resistant transgenic plants. J Gen Virol. 1999;80:1339–1346. doi: 10.1099/0022-1317-80-6-1339. [DOI] [PubMed] [Google Scholar]
- 9.Breyer WA, Matthews BW. A structural basis for processivity. Protein Sci. 2001;10:1699–1711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Von Hippel PH, Fairfield FR, Dolejsi MK. On the processivity of polymerases. Ann NY Acad Sci. 1994;726:118–131. doi: 10.1111/j.1749-6632.1994.tb52803.x. [DOI] [PubMed] [Google Scholar]
- 11.Baird HA, et al. Sequence determinants of breakpoint location during HIV-1 intersubtype recombination. Nucleic Acids Res. 2006;34:5203–5216. doi: 10.1093/nar/gkl669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galetto R, Giacomoni V, Véron M, Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J Biol Chem. 2006;281:2711–2720. doi: 10.1074/jbc.M505457200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Temin HM. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol. 1994;68:2409–2414. doi: 10.1128/jvi.68.4.2409-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. A review of recombination frequency in RNA viruses, focusing particularly on the use of phylogenetic methods to investigate this characteristic. [DOI] [PubMed] [Google Scholar]
- 15.Huang AS, Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 16.Roux L, Simon AE, Holland JJ. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazzarini RA, Keene JD, Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981;26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 18.Chetverin AB, Chetverina HV, Demidenko AA, Ugarov VI. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell. 1997;88:503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallei A, Pankraz A, Thiel HJ, Becher P. RNA recombination in vivo in the absence of viral replication. J Virol. 2004;78:6271–6281. doi: 10.1128/JVI.78.12.6271-6281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gmyl AP, et al. Nonreplicative RNA recombination in poliovirus. J Virol. 1999;73:8958–8965. doi: 10.1128/jvi.73.11.8958-8965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald SM, Patton JT. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011;19:136–144. doi: 10.1016/j.tim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nibert ML, Margraf RL, Coombs KM. Nonrandom segregation of parental alleles in reovirus reassortants. J Virol. 1996;70:7295–7300. doi: 10.1128/jvi.70.10.7295-7300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu WP, Geske SM, Hickey CM, Moyer JW. Tomato spotted wilt Tospovirus genome reassortment and genome segment-specific adaptation. Virology. 1998;244:186–194. doi: 10.1006/viro.1998.9131. [DOI] [PubMed] [Google Scholar]
- 24.Urquidi V, Bishop DH. Non-random reassortment between the tripartite RNA genomes of La Crosse and snowshoe hare viruses. J Gen Virol. 1992;73:2255–2265. doi: 10.1099/0022-1317-73-9-2255. [DOI] [PubMed] [Google Scholar]
- 25.Roossinck MJ. Symbiosis versus competition in plant virus evolution. Nature Rev Microbiol. 2005;3:917–924. doi: 10.1038/nrmicro1285. [DOI] [PubMed] [Google Scholar]
- 26.González-Jara P, Fraile A, Canto T, García-Arenal F. The multiplicity of infection of a plant virus varies during colonization of its eukaryotic host. J Virol. 2009;83:7487–7494. doi: 10.1128/JVI.00636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyashita S, Kishino H. Estimation of the size of genetic bottlenecks in cell-to-cell movement of Soilborne wheat mosaic virus and the possible role of the bottlenecks in speeding up selection of variations in trans-acting genes or elements. J Virol. 2010;84:1828–1837. doi: 10.1128/JVI.01890-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Arriaza J, Manrubia SC, Toja M, Domingo E, Escarmís C. Evolutionary transition toward defective RNAs that are infectious by complementation. J Virol. 2004;78:11678–11685. doi: 10.1128/JVI.78.21.11678-11685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond DA, Silberg JJ, Meyer MM, Wilke CO, Arnold FH. On the conservative nature of intragenic recombination. Proc Natl Acad Sci USA. 2005;102:5380–5385. doi: 10.1073/pnas.0500729102. An important paper exploring the relative impact of mutation and recombination on the functionality of proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voigt CA, Martinez C, Wang ZG, Mayo SL, Arnold FH. Protein building blocks preserved by recombination. Nature Struct Biol. 2002;9:553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- 31.Escriu F, Fraile A, García-Arenal F. Constraints to genetic exchange support gene coadaptation in a tripartite RNA virus. PLoS Pathog. 2007;3:e8. doi: 10.1371/journal.ppat.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pringle CR, Lees JF, Clark W, Elliott RM. Genome subunit reassortment among Bunyaviruses analysed by dot hybridization using molecularly cloned complementary DNA probes. Virology. 1984;135:244–256. doi: 10.1016/0042-6822(84)90134-x. [DOI] [PubMed] [Google Scholar]
- 33.Mansky LM. Retrovirus mutation rates and their role in genetic variation. J Gen Virol. 1998;79:1337–1345. doi: 10.1099/0022-1317-79-6-1337. [DOI] [PubMed] [Google Scholar]
- 34.Neher RA, Leitner T. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput Biol. 2010;6:e1000660. doi: 10.1371/journal.pcbi.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shriner D, Rodrigo AG, Nickle DC, Mullins JI. Pervasive genomic recombination of HIV-1 in vivo. Genetics. 2004;167:1573–1583. doi: 10.1534/genetics.103.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savolainen-Kopra C, Blomqvist S. Mechanisms of genetic variation in polioviruses. Rev Med Virol. 2010;20:358–371. doi: 10.1002/rmv.663. [DOI] [PubMed] [Google Scholar]
- 37.Lai MM, Pearlman S, Anderson LJ. In: Fields Virology. 5. Howley PM, Knipe DM, editors. Lippincott Williams and Wilkins; Philadelphia, USA: 2007. pp. 1305–1335. [Google Scholar]
- 38.Urbanowicz A, et al. Homologous crossovers among molecules of brome mosaic bromovirus RNA1 or RNA2 segments in vivo. J Virol. 2005;79:5732–5742. doi: 10.1128/JVI.79.9.5732-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibbs A, Ohshima K. Potyviruses and the digital revolution. Annu Rev Phytopathol. 2010;48:205–223. doi: 10.1146/annurev-phyto-073009-114404. [DOI] [PubMed] [Google Scholar]
- 40.Tomimura K, et al. Comparisons of the genetic structure of populations of Turnip mosaic virus in West and East Eurasia. Virology. 2004;330:408–423. doi: 10.1016/j.virol.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 41.Holmes EC, Worobey M, Rambaut A. Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol. 1999;16:405–409. doi: 10.1093/oxfordjournals.molbev.a026121. [DOI] [PubMed] [Google Scholar]
- 42.Taucher C, Berger A, Mandl CW. A trans-complementing recombination trap demonstrates a low propensity of flaviviruses for intermolecular recombination. J Virol. 2010;84:599–611. doi: 10.1128/JVI.01063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolou HJ, et al. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J Gen Virol. 2001;82:1283–1290. doi: 10.1099/0022-1317-82-6-1283. [DOI] [PubMed] [Google Scholar]
- 44.Uzcategui NY, et al. Molecular epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J Gen Virol. 2001;82:2945–2953. doi: 10.1099/0022-1317-82-12-2945. [DOI] [PubMed] [Google Scholar]
- 45.Reiter J, et al. Hepatitis C virus RNA recombination in cell culture. J Hepatol. 2011 Feb 17; doi: 10.1016/j.jhep.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 46.Chare ER, Gould EA, Holmes EC. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J Gen Virol. 2003;84:2691–2703. doi: 10.1099/vir.0.19277-0. A study that uses a comparative phylogenetic analysis to show the low rate of recombination in (−)ssRNA viruses. [DOI] [PubMed] [Google Scholar]
- 47.McCarthy AJ, Shaw MA, Goodman SJ. Pathogen evolution and disease emergence in carnivores. Proc Biol Sci. 2007;274:3165–3174. doi: 10.1098/rspb.2007.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boni MF, de Jong MD, van Doorn HR, Holmes EC. Guidelines for identifying homologous recombination events in influenza A virus. PLoS ONE. 2010;5:e10434. doi: 10.1371/journal.pone.0010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatchette TF, et al. Influenza A viruses in feral Canadian ducks: extensive reassortment in nature. J Gen Virol. 2004;85:2327–2337. doi: 10.1099/vir.0.79878-0. [DOI] [PubMed] [Google Scholar]
- 50.Lindstrom SE, Cox NJ, Klimov A. Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957–1972: evidence for genetic divergence and multiple reassortment events. Virology. 2004;328:101–119. doi: 10.1016/j.virol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Xu X, et al. Reassortment and evolution of current human influenza A and B viruses. Virus Res. 2004;103:55–60. doi: 10.1016/j.virusres.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Jetzt AE, et al. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung A, et al. Recombination: multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 54.Nemirov K, et al. Isolation and characterization of Dobrava hantavirus carried by the striped field mouse (Apodemus agrarius) in Estonia. J Gen Virol. 1999;80:371–379. doi: 10.1099/0022-1317-80-2-371. [DOI] [PubMed] [Google Scholar]
- 55.Vieth S, Torda AE, Asper M, Schmitz H, Gunther S. Sequence analysis of L RNA of Lassa virus. Virology. 2004;318:153–168. doi: 10.1016/j.virol.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Miranda GJ, Azzam O, Shirako Y. Comparison of nucleotide sequences between northern and southern philippine isolates of rice grassy stunt virus indicates occurrence of natural genetic reassortment. Virology. 2000;266:26–32. doi: 10.1006/viro.1999.0068. [DOI] [PubMed] [Google Scholar]
- 57.Rabadan R, Levine AJ, Krasnitz M. Non-random reassortment in human influenza A viruses. Influenza Other Respi Viruses. 2008;2:9–22. doi: 10.1111/j.1750-2659.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iturriza-Gómara M, Isherwood B, Desselberger U, Gray J. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J Virol. 2001;75:3696–3705. doi: 10.1128/JVI.75.8.3696-3705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe M, Nakagomi T, Koshimura Y, Nakagomi O. Direct evidence for genome segment reassortment between concurrently-circulating human rotavirus strains. Arch Virol. 2001;146:557–570. doi: 10.1007/s007050170162. [DOI] [PubMed] [Google Scholar]
- 60.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nature Rev Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 61.McDonald SM, et al. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog. 2009;5:e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silander OK, et al. Widespread genetic exchange among terrestrial bacteriophages. Proc Natl Acad Sci USA. 2005;102:19009–19014. doi: 10.1073/pnas.0503074102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rice WR. Experimental tests of the adaptive significance of sexual recombination. Nature Rev Genet. 2002;3:241–251. doi: 10.1038/nrg760. [DOI] [PubMed] [Google Scholar]
- 64.Burt A. Perspective: sex, recombination, and the efficacy of selection – was Weismann right? Evolution. 2000;54:337–351. doi: 10.1111/j.0014-3820.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 65.Gu Z, Gao Q, Faust EA, Wainberg MA. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J Gen Virol. 1995;76:2601–2605. doi: 10.1099/0022-1317-76-10-2601. [DOI] [PubMed] [Google Scholar]
- 66.Kellam P, Larder BA. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J Virol. 1995;69:669–674. doi: 10.1128/jvi.69.2.669-674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moutouh L, Corbeil J, Richman DD. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci USA. 1996;93:6106–6111. doi: 10.1073/pnas.93.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yusa K, Kavlick MF, Kosalaraksa P, Mitsuya H. HIV-1 acquires resistance to two classes of antiviral drugs through homologous recombination. Antiviral Res. 1997;36:179–189. doi: 10.1016/s0166-3542(97)00053-3. [DOI] [PubMed] [Google Scholar]
- 69.Kalinina O, Norder H, Mukomolov S, Magnius LO. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg J Virol. 2002;76:4034–4043. doi: 10.1128/JVI.76.8.4034-4043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noppornpanth S, et al. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol. 2006;80:7569–7577. doi: 10.1128/JVI.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sentandreu V, et al. Evidence of recombination in intrapatient populations of hepatitis C virus. PLoS ONE. 2008;3:e3239. doi: 10.1371/journal.pone.0003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miralles R, Gerrish PJ, Moya A, Elena SF. Clonal interference and the evolution of RNA viruses. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. An important experimental demonstration of clonal interference in RNA viruses. [DOI] [PubMed] [Google Scholar]
- 74.Pepin KM, Wichman HA. Experimental evolution and genome sequencing reveal variation in levels of clonal interference in large populations of bacteriophage phiX174. BMC Evol Biol. 2008;8:85. doi: 10.1186/1471-2148-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poon A, Chao L. Drift increases the advantage of sex in RNA bacteriophage phi6. Genetics. 2004;166:19–24. doi: 10.1534/genetics.166.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 77.Chao L. Fitness of RNA virus decreased by Muller’s ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. A classic demonstration of the occurrence of Muller’s ratchet in an experimental RNA virus population. [DOI] [PubMed] [Google Scholar]
- 78.Chao L. In: The Evolutionary Biology of Viruses. Morse SS, editor. Raven; New York: 1994. pp. 233–250. [Google Scholar]
- 79.Chao L, Tran TT, Matthews C. Muller’s ratchet and the advantage of sex in the RNA virus phi-6. Evolution. 1992;46:289–299. doi: 10.1111/j.1558-5646.1992.tb02038.x. [DOI] [PubMed] [Google Scholar]
- 80.Chao L, Tran TT. The advantage of sex in the RNA virus phi6. Genetics. 1997;147:953–959. doi: 10.1093/genetics/147.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lázaro E, Escarmís C, Pérez-Mercader J, Manrubia SC, Domingo E. Resistance of virus to extinction on bottleneck passages: study of a decaying and fluctuating pattern of fitness loss. Proc Natl Acad Sci USA. 2003;100:10830–10835. doi: 10.1073/pnas.1332668100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keightley PD, Eyre-Walker A. Deleterious mutations and the evolution of sex. Science. 2000;290:331–333. doi: 10.1126/science.290.5490.331. [DOI] [PubMed] [Google Scholar]
- 83.Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–440. doi: 10.1038/336435a0. A seminal report outlining the mutational deterministic hypothesis for the evolution of sexual reproduction. [DOI] [PubMed] [Google Scholar]
- 84.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nature Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 85.Elena SF. Little evidence for synergism among deleterious mutations in a nonsegmented RNA virus. J Mol Evol. 1999;49:703–707. doi: 10.1007/pl00000082. [DOI] [PubMed] [Google Scholar]
- 86.Pybus OG, et al. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 87.Bonhoeffer S, Chappey C, Parkin NT, Whitcomb JM, Petropoulos CJ. Evidence for positive epistasis in HIV-1. Science. 2004;306:1547–1550. doi: 10.1126/science.1101786. An important study showing that positive (antagonistic) epistasis occurs in HIV and, hence, that recombination is unlikely to be selected as a way of purging deleterious mutations. [DOI] [PubMed] [Google Scholar]
- 88.Burch CL, Turner PE, Hanley KA. Patterns of epistasis in RNA viruses: a review of the evidence from vaccine design. J Evol Biol. 2003;16:1223–1235. doi: 10.1046/j.1420-9101.2003.00632.x. [DOI] [PubMed] [Google Scholar]
- 89.Sanjuan R, Moya A, Elena SF. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci USA. 2004;101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shapiro B, Rambaut A, Pybus OG, Holmes EC. A phylogenetic method for detecting positive epistasis in gene sequences and its application to RNA virus evolution. Mol Biol Evol. 2006;23:1724–1730. doi: 10.1093/molbev/msl037. [DOI] [PubMed] [Google Scholar]
- 91.Elena SF, Carrasco P, Daros JA, Sanjuan R. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 2006;7:168–173. doi: 10.1038/sj.embor.7400636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner PE, Chao L. Sex and the evolution of intrahost competition in RNA virus phi6. Genetics. 1998;150:523–532. doi: 10.1093/genetics/150.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fulton RW. Practices and precautions in the use of cross protection for plant-virus disease control. Annu Rev Phytopathol. 1986;24:67–81. [Google Scholar]
- 94.Nethe M, Berkhout B, van der Kuyl AC. Retroviral superinfection resistance. Retrovirology. 2005;2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J Virol. 2005;79:3231–3242. doi: 10.1128/JVI.79.6.3231-3242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Adams RH, Brown DT. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J Virol. 1985;54:351–357. doi: 10.1128/jvi.54.2.351-357.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J Virol. 1997;71:7119–7123. doi: 10.1128/jvi.71.9.7119-7123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Michod RE, Bernstein H, Nedelcu AM. Adaptive value of sex in microbial pathogens. Infect Genet Evol. 2008;8:267–285. doi: 10.1016/j.meegid.2008.01.002. An article that outlines the repair hypothesis for the evolution of sexual reproduction and applies it to microbial populations, including RNA viruses. [DOI] [PubMed] [Google Scholar]
- 99.Coffin JM. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 100.Xu H, Boeke JD. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:8553–8557. doi: 10.1073/pnas.84.23.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu WS, Temin HM. Effect of gamma radiation on retroviral recombination. J Virol. 1992;66:4457–4463. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Novella IS, Ball LA, Wertz GW. Fitness analyses of vesicular stomatitis strains with rearranged genomes reveal replicative disadvantages. J Virol. 2004;78:9837–9841. doi: 10.1128/JVI.78.18.9837-9841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spann KM, Collins PL, Teng MN. Genetic recombination during coinfection of two mutants of human respiratory syncytial virus. J Virol. 2003;77:11201–11211. doi: 10.1128/JVI.77.20.11201-11211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Archer AM, Rico-Hesse R. High genetic divergence and recombination in Arenaviruses from the Americas. Virology. 2002;304:274–281. doi: 10.1006/viro.2002.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Charrel RN, et al. Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem Biophys Res Commun. 2002;296:1118–1124. doi: 10.1016/s0006-291x(02)02053-3. [DOI] [PubMed] [Google Scholar]
- 106.Wittmann TJ, et al. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc Natl Acad Sci USA. 2007;104:17123–17127. doi: 10.1073/pnas.0704076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kondrashov AS, Crow JF. Haploidy or diploidy: which is better? Nature. 1991;351:314–315. doi: 10.1038/351314a0. [DOI] [PubMed] [Google Scholar]
- 110.Otto SP, Goldstein DB. Recombination and the evolution of diploidy. Genetics. 1992;131:745–751. doi: 10.1093/genetics/131.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Perrot V, Richerd S, Valero M. Transition from haploidy to diploidy. Nature. 1991;351:315–317. doi: 10.1038/351315a0. [DOI] [PubMed] [Google Scholar]
- 112.Froissart R, et al. Recombination every day: abundant recombination in a virus during a single multi-cellular host infection. PLoS Biol. 2005;3:e89. doi: 10.1371/journal.pbio.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hudson RR. Estimating the recombination parameter of a finite population model without selection. Genet Res. 1987;50:245–250. doi: 10.1017/s0016672300023776. [DOI] [PubMed] [Google Scholar]
- 114.Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Simon-Loriere E, et al. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 2009;5:e1000418. doi: 10.1371/journal.ppat.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nikolaitchik OA, Galli A, Moore MD, Pathak VK, Hu WS. Multiple barriers to recombination between divergent HIV-1 variants revealed by a dual-marker recombination assay. J Mol Biol. 2011;407:521–531. doi: 10.1016/j.jmb.2011.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Motomura K, Chen J, Hu WS. Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. J Virol. 2008;82:1923–1933. doi: 10.1128/JVI.01937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marsh GA, Rabadan R, Levine AJ, Palese P. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J Virol. 2008;82:2295–2304. doi: 10.1128/JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Simon-Loriere E, Martin DP, Weeks KM, Negroni M. RNA structures facilitate recombination-mediated gene swapping in HIV-1. J Virol. 2010;84:12675–12682. doi: 10.1128/JVI.01302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weaver SC. Evolutionary influences in arboviral disease. Curr Top Microbiol Immunol. 2006;299:285–314. doi: 10.1007/3-540-26397-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jackwood MW, et al. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martin GS. The road to Src. Oncogene. 2004;23:7910–7917. doi: 10.1038/sj.onc.1208077. [DOI] [PubMed] [Google Scholar]