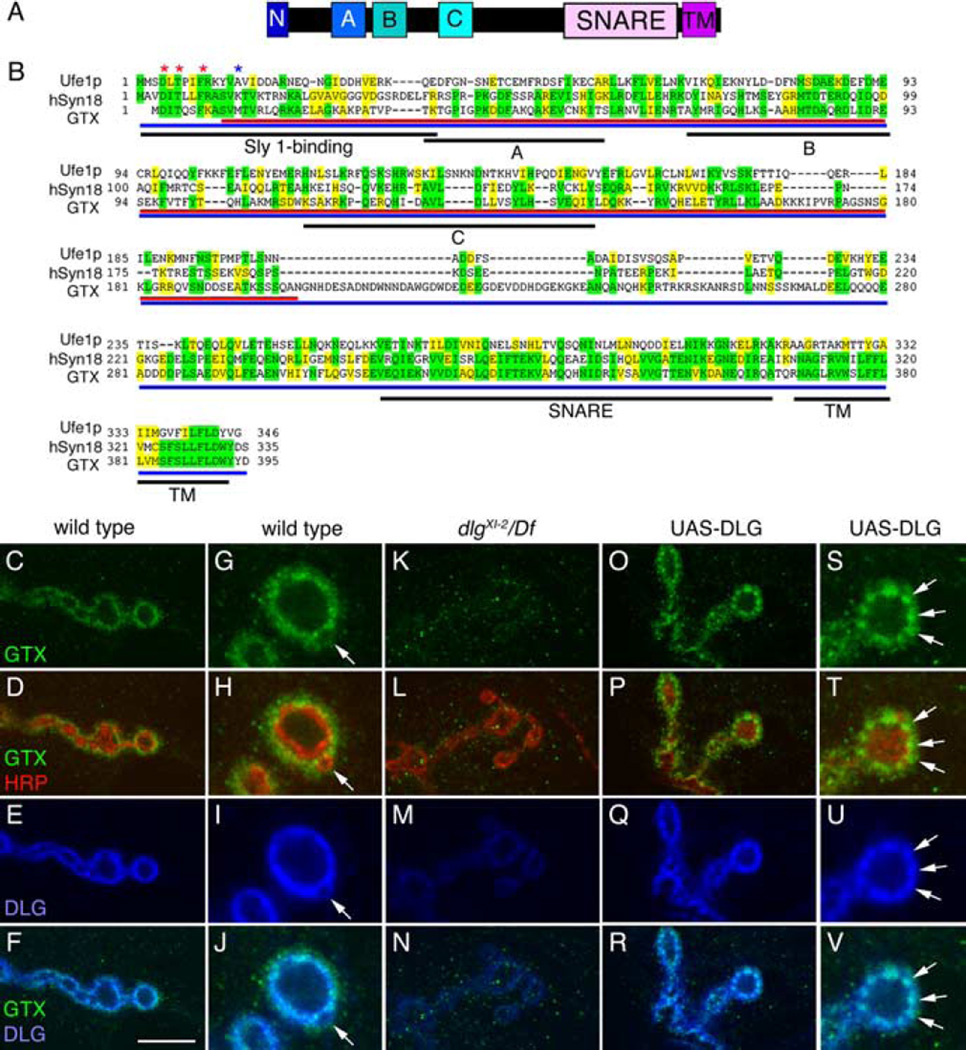
Figure 1.
GTX protein structure and synaptic localization. A, Organization of GTX into protein domains. (N, N-terminal homologous Sly1 binding site; A, B, C, predicted Ha, Hb, and Hc domains; SNARE, coiled-coil SNARE domain; TM, transmembrane domain). B, Protein alignment of yeast Ufe1p, human Syntaxin 18, and GTX. Asterisks represent amino acids required for Sly1 binding (red, conserved; blue, not conserved). Regions of identity are highlighted in yellow, whereas conserved regions are marked in green. Black underlining indicates different protein domains. Red and blue underlining shows the GTX protein regions used to generate the anti-GTX antisera. C–V, Single confocal slices of third instar larval NMJs labeled with anti-GTX (green), anti-HRP (red), and anti-DLG (blue), shown at low (C–F, K–R) and high (G–J, S–V) magnification in wild type (C–J), dlgXI−2/Df(dlg) (K–N), and UAS-DLG (O–V) overexpressed using the BG487 Gal4. The arrow in G–J points to a budding bouton. Arrows in S–V point to accumulations of DLG, which are accompanied by accumulations of GTX. Scale bar: C–F, K–N, O–R, 7 µm; G–J, S–V, 3 µm.