Abstract
Chordoma and chondrosarcoma are malignant bone tumors characterized by the abundant production of extracellular matrix. The resistance of these tumors to conventional therapeutic modalities has prompted us to delineate the gene expression profile of these two tumor types, with the expectation to identify potential molecular therapeutic targets. Furthermore the transcriptional profile of chordomas and chrondrosarcomas was compared to a wide variety of sarcomas as well as to that of normal tissues of similar lineage, to determine whether they express unique gene signatures among other tumors of mesenchymal origin, and to identify changes associated with malignant transformation. A HG-U133A Affymetrix Chip platform was used to determine the gene expression signature in 6 chordoma and 14 chondrosarcoma lesions. Validation of selected genes was performed by qPCR and immunohistochemistry (IHC) on an extended subset of tumors. By unsupervised clustering, chordoma and chondrosarcoma tumors grouped together in a genomic cluster distinct from that of other sarcoma types. They shared overexpression of many extracellular matrix genes including aggrecan, type II & X collagen, fibronectin, matrillin 3, high molecular weight-melanoma associated antigen (HMW-MAA), matrix metalloproteinase MMP-9, and MMP-19. In contrast, T Brachyury and CD24 were selectively expressed in chordomas, as were Keratin 8,13,15,18 and 19. Chondrosarcomas are distinguished by high expression of type IX and XI collagen. Because of its potential usefulness as a target for immunotherapy, the expression of HMW-MAA was analyzed by IHC and was detected in 62% of chordomas and 48% of chondrosarcomas, respectively. Furthermore, western blotting analysis showed that HMW-MAA synthesized by chordoma cell lines has a structure similar to that of the antigen synthesized by melanoma cells. In conclusion, chordomas and chondrosarcomas share a similar gene expression profile of up-regulated extracellular matrix genes. HMW-MAA represents a potential useful target to apply immunotherapy to these tumors.
Keywords: Chordoma, Chondrosarcoma, Gene expression, Extracellular matrix, HMW-MAA
Introduction
Chordomas and chondrosarcomas are rare malignant bone tumors which share several characteristics, but also display distinctive features. First, both tumors produce abundant extracellular matrix which contributes to their histologic identification. Second, both tumors express several proteins known to be important in cartilage matrix production including aggrecan, type II collagen, cartilage oligomeric matrix protein and SOX-9. Third, some chordomas have a chondroid component which is histologically similar to low grade chondrosarcomas [19, 35]. As a result, the histopathological distinction between these chordomas and chondrosarcomas may be difficult, particularly when needle biopsies are utilized for diagnostic purposes. Fourth, both tumors have similar rates of metastasis, since conventional chordomas and high grade chondrosarcomas metastasize in 30–40% of cases [3, 6, 13, 14, 39]. Lastly, for both tumors surgical excision is the main therapy modality, since neither of them can be effectively treated with conventional chemotherapies and/or photon radiation [13, 29, 34]. However, unlike chondrosarcomas, which occur in the appendicular as well as in the axial skeleton, chordomas arise predominantly in the sacrococcygeal and spheno-occipital regions of the axial skeleton [4, 16, 17, 23]. Furthermore, utilizing gene expression microarrays and immunohistochemical staining of formalin-fixed tissue sections with a polyclonal antibody, Vujovic et al. [43] demonstrated antigenic differences between chordoma and chondrosarcoma. T Brachyury, a transcription factor known to be involved in notochord development, was found to be expressed in all 53 chordoma lesions tested, but was not detectable in over 300 malignant tumors, including 163 chondroid tumors [43]. The differential expression of T Brachyury in chordomas and chondrosarcomas has lead to the suggestion that this molecule is a sensitive and specific marker to distinguish chordomas, including chondroid chordomas, from chondrosarcomas [18, 43]. These findings have been extended to soft tissue chordomas by Tirabosco et al. [41] who have confirmed the lack of expression of T Brachyury in carcinomas, lymphomas and sarcomas. In contrast, Palena et al. [31] have described the expression of T Brachyury in various types of carcinomas.
In this study, we have analyzed the gene profile of chordomas and chondrosarcomas to determine whether molecules shown to be appropriate targets to apply immunotherapy are expressed in these tumors. Furthermore we have compared the gene profile of chordomas and chondrosarcomas to that of a wide variety of sarcomas to determine whether they express a unique gene signature among tumors of mesenchymal origin. Lastly, to identify transcriptional changes associated with malignant transformation, we have compared the gene profile of chordomas and chondrosarcomas to that of normal tissues of similar lineage, i.e., nucleus pulposus and articular cartilage, respectively. The latter is similar to chondrosarcoma in its hyaline matrix production. Nucleus pulposus was used, since it might contain notochordal cells from which chordoma is thought to arise.
Materials and methods
Cell lines
The human primary chondrosarcoma cell lines JJ and KC [8], the human primary chordoma cell line, the human metastatic chordoma cell line [27] and the human melanoma cell line Colo 38 were grown at 37°C in a 5% CO2 atmosphere in RPMI 1640 medium supplemented with 10% fetal calf serum (BioWhittaker, Walkersville, MD).
Tumor samples
Patients with a pathologic diagnosis of chordoma and chondrosarcoma were identified from the Pathology Department files of Memorial Sloan-Kettering Cancer Center. Those who underwent an in-house surgical resection and had snap-frozen tumor available were selected for the study. The pathologic diagnosis as well as the histologic grade were re-confirmed in all cases by review of the resected lesion and corroborated by the clinical and radiographic findings. Inclusion criteria for chondrosarcoma patients were a diagnosis of primary chondrosarcoma in the long bones or axial skeleton, conventional histology, predominantly from the low to intermediate grade category, as potentially having less secondary genetic alterations. Secondary chondrosarcoma cases or tumors having a non-conventional morphology, such as mesenchymal, clear cell, etc., were excluded. Only chordomas with a conventional morphology were included in the study, dedifferentiated and chondroid chordoma subtypes were excluded. Six chordomas and 14 chondrosarcomas matched these inclusion criteria. The chordoma patients included five males and one female, with an average age at diagnosis of 66 years (range 54–76). The chordoma tumor location included four sacral, one clivus and one cervical spine, and they all showed a conventional-type morphology. The chondrosarcoma patients had an average age at diagnosis of 52 years (range 33–75) and their tumors were located in the long bones (proximal humerus, 4; proximal femur, 3; distal femur, 2) or in the flat bones of the axial skeleton (rib, 3; pelvis, 2). The histologic grade of these tumors included five grade I, seven grade II, and two grade III tumors. The grade I tumors were classified as stage IA in five cases and stage IIB in other two. All the remaining higher grade tumors were stage IIB.
Nucleus pulposus and articular cartilage were included as normal controls for the gene expression analysis of chordomas and chondrosarcomas, respectively. The nucleus pulposus and facet articular cartilage samples were procured from two male patients (ages 52 and 60 years) undergoing fusion of their lumbar spine for degenerative spondylosis. In addition, a heterogeneous group of 45 soft tissue sarcomas, of 7 histologic types including 6 fibrosarcomas, 6 leiomyosarcomas, 4 synovial sarcomas, 5 clear cell sarcomas, 4 gastrointestinal stromal tumors, 12 liposarcoma and 8 malignant fibrous histiocytomas, with previously available gene expression data was used as a control group [37]. This study was approved by the Institutional Review Board.
U133A Chip Affymetrix microarray and data analysis
A total of 14 chondrosarcomas and 6 chordomas revealed good-quality RNA and were utilized for gene expression analysis as previously described [1]. The images were quantified using a GCOS1.1 (GeneChip Operating System, Affymetrix) with the default parameters for the statistical algorithm and all probe set scaling with a target intensity of 500 to account for differences in the global chip intensity. The expression values were transformed using the logarithm base 2 for all subsequent analyses. The data analysis was performed using two methods. The first one analyzed the log of the normalized expression data using the LIMMA method from the Bioconductor package. This method uses a modified t statistic, which adds a correction term to the sample variances. To control for the multiple testing problem, the false discovery rate (FDR) method was used and the list was cut off at an FDR of 1e10−04. The gene lists obtained for each individual analysis were cross-referenced against both the published literature and the gene ontology consortium database (http://www.geneontology.org/) using NetAffx (http://www.affymetrix.com). Hierarchical clustering was performed using the Pearson correlation metric and average linkage. A filter was applied to remove any genes scored absent in over 75% of the samples. To assess the robustness of the clustering result, bootstrap resampling was done [46]. A parametric resampling method was used to simulate noise in the data. A total of 1,000 bootstrap datasets were computed and each replica of the data was clustered. The 1,000 trees were then combined using a majority rule algorithm [46] to compute the consensus tree. Each node was scored by how many times it appeared in the 1,000 bootstrap trees, with a high value indicating a robust subcluster. The second method of data analysis used the Affymetrix Genespring 7.2 software. For identifying differentially expressed genes, the 22,000 genes were filtered for flags and expression values and a gene list that selected genes with ≥2-fold change between the groups was identified. Using these two methods we performed the following analyses: chordoma versus soft tissue sarcoma, chondrosarcoma versus soft tissue sarcoma, chordoma versus lumbar nucleus pulposus, chondrosarcoma versus lumbar facet articular cartilage, and an unsupervised clustering analysis of all malignant tumors (chordoma, chondrosarcoma and soft tissue sarcoma). A Venn diagram function was used to identify the genes that were in common between the chordoma group and the chondrosarcoma group when compared with the same set of soft tissue sarcomas.
Real-time PCR
Quantitative gene expression analysis was performed for CD24, HMW-MAA, T Brachyury and type IX collagen using the Thermosript RT-PCR system (Invitrogen Life Technologies), as previously described [2]. Genes chosen for validation were based on their preferential expression in chordoma (CD24 and T Brachyury) or chondrosarcoma (type IX Collagen). HMW-MAA was chosen due to its expression in both tumor types, as well as to its potential use as a target for immunotherapy. Statistical analysis was carried out on SPSS software (SPSS, version 12, Chicago, IL). The Whitney Mann test was utilized and a P value of less than 0.05 was considered statistically significant.
Monoclonal antibodies
The HMW-MAA-specific mAb 116, 149.53, 225.28, 724, 763.74, TP41.2, TP43, TP61.5, TP108, TP109, VF18.176 VT1.7, VT5-1, VT67.5, VT68.2 and VT80.12 [9] and the anti-idiotypic (anti-id) mAb MK2-23 [20] were developed and characterized as described. mAb were purified from ascitic fluid by sequential precipitation with ammonium sulphate and caprylic acid [40]. The purity of mAb preparations was monitored by SDS-PAGE; their activity was monitored by testing with HMW-MAA bearing melanoma cells in ELISA.
Immunohistochemistry
Three Tissue MicroArrays (TMA) were constructed using an automated arrayer (ATA-27, Beecher Instruments, Sun Prarie, WI), to include 21 conventional chordoma and 84 chondrosarcoma lesions. From each sample triplicate cores were used, each measuring 0.6 mm in diameter. Each slide was incubated for 3 h at 37°C in a closed humid chamber with the pool of the HMW-MAA-specific mAb 763.74, VF1-TP41.2 and VT80.12 (20 μg/0.1 ml PBS). The indicated pool of mAb was used, since in preliminary experiments it was found to be the most sensitive to detect HMW-MAA in formalin-fixed, paraffin-embedded tissue sections. Following a 30-min incubation at room temperature (RT) with methanol containing 0.3% hydrogen peroxide to block endogenous peroxidase activity, samples were treated with Hyaluronidase (1% 1 × PBS, pH 5.5) and washed in PBS, pH 7.4. Samples were subjected to antigen retrieval using 10 mM Tris buffer, pH 10.0 containing 0.5% Tween-20 and microwave heating for 30 min. Non-specific binding was inhibited with protein block (Dako, Carpinteria, CA) and 0.5% bovine serum albumin for 30 min at RT in a humid chamber. PBS, normal rabbit IgG, or normal goat IgG (Sigma Chemical, St. Louis, MO) were used instead of the primary antibodies to monitor the specificity of the staining. Samples were washed in PBS and incubated with the biotinylated secondary antibody for 1 h at RT. The avidin–biotin–peroxidase complex was then added and incubation was continued for 30 min at RT. Antibody localization was determined with the diaminobenzidine reaction for 10–20 min at RT. Immunohistochemical staining was evaluated independently by two pathologists (CRA, NPA) and scored as present or absent.
Cell staining and flow cytometric analysis
Staining of cells with HMW-MAA-specific mAb 225.28 and flow cytometric analysis utilizing a FACS-can™ flow cytometer (BD Biosciences, San Jose, CS) were performed as described [22]. The anti-id mAb MK-23 was used as a specificity control. Data were analyzed utilizing the Cell-Quest software (BD Biosciences, San Jose, CA).
Western blotting analysis
Western blotting analysis was performed utilizing cell line lysates and the HMW-MAA-specific mAb 763.74 as described [20]. The HMW-MAA bearing melanoma cell line Colo 38 and the anti-id mAb MK2-23 were used as controls.
Results
Up-regulation of extracellular matrix genes in both chordomas and chondrosarcomas
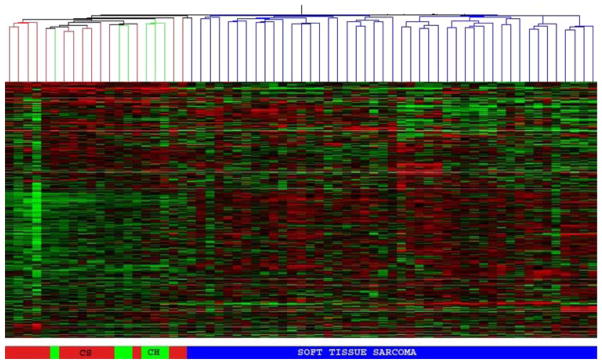
The unsupervised hierarchical clustering analysis, using both the bootstrap clustering and the GeneSpring software, showed that chordoma and chondrosarcoma samples clustered together as a single genomic group, distinct from all the other soft tissue sarcoma samples (Fig. 1). The gene lists obtained using both statistical methods were compared and showed a similar set of differentially expressed genes. Gene lists from the First method were used for further analysis and identification of genes. Most of the genes found to be overexpressed in both chordoma and chondrosarcoma, relative to soft tissue sarcomas, are involved in the synthesis or regulation of extracellular matrix. Some of the top-ranked genes are involved in the synthesis of major constituents of the extracellular matrix (type II collagen, aggrecan, type X collagen, matrillin 3), matrix metabolism and degradation [Matrix metalloproteinase (MMP) 9 and 19], and cell-matrix interactions (fibronectin, integrins, HMW-MAA and ADAM28) (Fig. 2; Table 1).
Fig. 1.
Unsupervised cluster analysis of chondrosarcoma (red), chordoma (green) and soft tissue sarcoma (blue). Chondrosarcoma and chordoma samples form a separate genomic group, readily delineated from soft tissue sarcoma
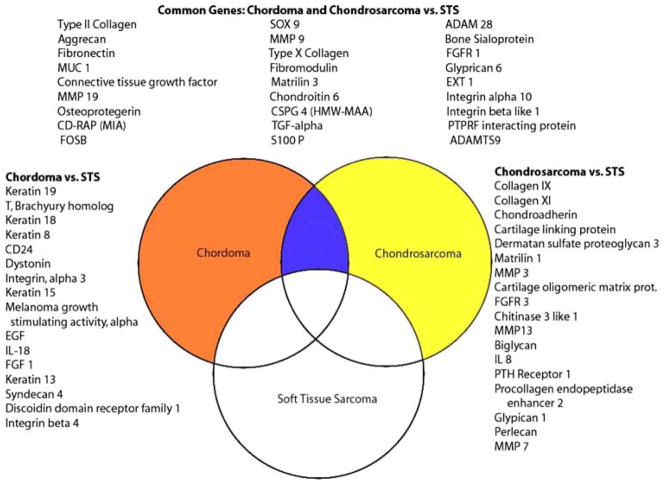
Fig. 2.
This Venn diagram demonstrates genes that show >2-fold change in chordoma (orange circle) and chondrosarcoma (yellow circle) relative to soft tissue sarcomas (white circle). The list on the left demonstrates selected genes that are up-regulated in chordoma relative to soft tissue sarcoma (STS). The list on the right demonstrates selected genes that are up-regulated in chondrosarcoma. The three columns at the top represent selected genes that are up-regulated in both chordoma and chondrosarcoma relative to soft tissue sarcoma
Table 1.
Genes differentially expressed in chordoma and chondrosarcoma tumors
| Gene symbol | Gene title | P value | Fold change | Chromosomal location | Gene ontology Biological process |
|---|---|---|---|---|---|
| Genes expressed in both chordoma and chondrosarcoma samples | |||||
| COL2A1 | Collagen, type II, alpha 1 | 4.77E-08 | 65.61 | 12q13.11 | Skeletal development |
| AGC1 | Aggrecan 1 | 1.22E-11 | 43.57 | 15q26.1 | Cell adhesion |
| FN1 | Fibronectin 1 | 6.4E-08 | 31.35 | 2q34 | Cell adhesion |
| MUC1 | Mucin 1, transmembrane | 1.04E-06 | 19.84 | 1q21 | Extracellular matrix |
| CTGF | Connective tissue growth factor | 0.00013 | 11.65 | 6q23.1 | Regulation of cell growth |
| HMW-MAA | High molecular weight melanoma associated antigen | 1.3E-05 | 9.54 | 15q23 | Cell motility |
| S100P | S100 calcium binding protein P | 3.82E-05 | 22.95 | 4p16 | Regulation of cell cycle |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 9.25E-06 | 19.41 | 19q13.32 | Regulation of cell cycle |
| MMP9 | Matrix metalloproteinase 9 | 7.72E-05 | 8.48 | 20q11.2 | Peptidoglycan metabolism |
| MMP19 | Matrix metalloproteinase 19 | 0.000155 | 10.87 | 12q14 | Peptidoglycan metabolism |
| ITGBL1 | Integrin, beta-like 1 | 0.000698 | 10.23 | 13q33 | Cell-matrix adhesion |
| SOX9 | SRY (sex determining region Y)-box 9 | 0.000194 | 7.07 | 17q24.3 | Skeletal development |
| MATN3 | Matrilin 3 | 0.000777 | 5.75 | 2p24-p23 | Skeletal development |
| FMOD | Fibromodulin | 3.04E-05 | 6.97 | 1q32 | Transforming growth factor beta receptor complex assembly |
| ADAM28 | A disintegrin and metalloproteinase domain 28 | 0.000499 | 3.83 | 8p21.2 | Proteolysis and peptidolysis |
| CHST3 | Carbohydrate (chondroitin 6) sulfotransferase 3 | 8.23E-07 | 6.27 | 10q22.1 | Chondroitin sulfate biosynthesis |
| Genes expressed specifically in chordomas | |||||
| KRT19 | Keratin 19 | 6.39E-16 | 1070.53 | 17q21.2 | Epidermis development |
| T | T, brachyury | 2.68E-19 | 79.79 | 6q27 | Mesoderm development |
| KRT18 | Keratin 18 | 6.96E-11 | 51.70 | 12q13 | Morphogenesis |
| KRT8 | Keratin 8 | 5.07E-15 | 29.70 | 12q13 | Cytoskeleton organization and biogenesis |
| ITGA3 | Integrin, alpha 3 | 1.84E-06 | 16.56 | 17q21.33 | Cell-matrix adhesion |
| KRT15 | Keratin 15 | 1.31E-10 | 15.94 | 17q21.2 | Epidermis development |
| CD24 | CD24 antigen | 0.000661 | 26.53 | 6q21 | Humoral immune response |
| DST | Dystonin | 1.52E-08 | 23.38 | 6p12-p11 | Cytoskeleton organization |
| EGF | Epidermal growth factor | 1.39E-06 | 10.57 | 4q25 | Positive regulation of cell proliferation |
| KRT13 | Keratin 13 | 9.26E-05 | 7.86 | 17q12-q21.2 | Epidermis development |
| SDC4 | Syndecan 4 | 0.000207 | 4.36 | 20q12 | Intracellular signaling |
| DDR1 | Discoidin domain receptor family, member 1 | 1.36E-07 | 4.11 | 6p21.3 | Transmembrane receptor protein tyrosine kinase signaling pathway |
| Genes expressed specifically in chondrosarcomas | |||||
| COL11A2 | Collagen, type XI, alpha 2 | 1.38E-23 | 261.31 | 6p21.3 | Skeletal development |
| COL9A1 | Collagen, type IX, alpha 1 | 1.91E-22 | 110.11 | 6q12-q14 | Phosphate transport |
| CHAD | Chondroadherin | 5.77E-20 | 31.16 | 17q21.33 | Regulation of cell growth |
| MMP3 | Matrix metalloproteinase 3 | 1.36E-14 | 10.53 | 11q22.3 | Collagen catabolism |
| COMP | Cartilage oligomeric matrix protein | 4.30E-15 | 10.48 | 19p13.1 | Extracellular matrix |
| HAPLN1 | Cartilage linking protein 1 | 2.73E-17 | 20.35 | 5q14.3 | Cell adhesion |
| MATN1 | Matrilin 1, cartilage matrix protein | 1.14E-09 | 16.16 | 1p35 | Cartilage condensation |
| MMP13 | Matrix metalloproteinase 13 | 2.47E-17 | 4.83 | 11q22.3 | Extracellular matrix |
| PTHR1 | Parathyroid hormone receptor 1 | 0.000251 | 3.53 | 3p22-p21.1 | Skeletal development |
Up-regulation of CD24, Keratin and T-Brachyury genes in chordoma samples and of collagen IX and XI genes in chondrosarcoma samples relative to soft tissue sarcomas
Chordoma and chondrosarcoma gene expression was also compared separately to the soft tissue sarcoma group. A total of 607 were noted to have at least 2-fold increased expression in chordoma samples relative to soft tissue sarcoma tumors. Genes that were up-regulated in chordomas included: CD24, epidermal growth factor, keratin 8, 13, 15, 18 and 19, and T Brachyury (Fig. 2; Table 1).
Similarly, the expression of 366 genes was increased at least 2-fold in chondrosarcoma samples relative to soft tissue sarcomas. The up-regulated genes included: collagen IX, collagen XI, dermatan sulfate proteoglycan 3, fibroblast growth factor receptor 3, parathyroid hormone receptor 1 (PTHR-1), and MMP 3, 7, and 13 (Fig. 2).
Overexpression of Insulin Growth Factor genes and components of the AP-1 transcription factor in chondrosarcoma relative to articular cartilage
The expression of insulin like growth factor 2 (IGF-2) was 69-fold higher in the chondrosarcoma samples relative to the articular cartilage from the lumbar spine. In addition, the expression of IGF-1 was increased 6-fold and that of several important IGF-binding protein genes (IGF-BP 2,3,4,6 and 7) was also overexpressed.
Components of the AP-1 transcription factor were found to be highly over expressed in chondrosarcomas. FOSB was expressed 108-fold relative to articular cartilage. FOSB was also overexpressed in chondrosarcomas relative to the soft tissue sarcomas. In addition to FOSB, c-jun and c-fos displayed a 6-fold higher expression in chondrosarcomas.
Many genes important to cartilage development and extracellular matrix were overexpressed in chondrosarcomas relative to soft tissue sarcomas as well as to articular cartilage (Fig. 2). These genes included: SOX 9, biglycan, dermatan sulfate proteoglycan, matrilin 1 and 3, integrin beta like 1, MMP7, collagen types IV, VI, VII, IX and XI, cartilage linking protein, fibronectin, EXT1 and FGFR3.
Overexpression of components of the AP-1 transcription factor and the epidermal growth factor receptor pathway in chordoma relative to nucleus pulposus
FOSB was one of the top ranked genes (FC, 114) showing overexpression in chordoma compared to nucleus pulposus. In addition, c-Fos (FC, fold change, 14) and c-jun (FC, 9) were highly expressed in chordomas. Both the epidermal growth factor receptor (EGFR) and the EGF ligand genes showed overexpression in chordoma relative to nucleus pulposus (FC of 6.6 and 4.9, respectively).
Many genes found to be up-regulated in chordomas relative to soft tissue sarcomas were also overexpressed compared to the nucleus pulposus (Fig. 2). Among them, the top ranked genes in both analyses were Keratin 19 (FC, 800), T Brachury (FC, 110) and CD24 (FC, 56). Several other genes were found to be overexpressed relative to both soft tissue sarcomas and nucleus pulposus including: Keratin 8, 13, 15, 17 and 18, MMP 9 and 19, SOX 9, Matrilin 3, TGF-α, S100 P, EXT1, integrin β like 1, integrin α 3, IL18 and discoidin domain receptor family (Fig. 2).
Real-time PCR validation of gene expression data was performed for four of the key-genes up-regulated in either or both chordoma and chondrosarcoma groups, including: HMW-MAA, CD24, T Brachyury and type IX Collagen (Table 2). Quantitative PCR confirmed the microarray results of overexpression of CD24 and T Brachyury genes in chordomas, relative to both chondrosarcoma and soft tissue sarcoma lesions, while Type IX collagen expression was validated by Real-Time PCR to be higher in chondrosarcoma than in chordoma and soft tissue sarcoma lesions (Table 2).
Table 2.
Validation by quantitative RT-PCR
| Gene designation | Groups compared | Fold change (RT-PCR) | Fold change (Microarray) | P value* | |
|---|---|---|---|---|---|
| COL9A1 | Collagen, type IX, alpha 1 | CS > chordoma | 407.5 | 118 | 0.03 |
| COL9A1 | Collagen, type IX, alpha 1 | CS > STS | 1549.6 | 93.2 | 0.03 |
| HMW-MAA | CS > STS | 3.87 | 3.87 | 0.03 | |
| HMW-MAA | Chordoma > STS | 7.7 | 9.5 | 0.009 | |
| HMW-MAA | Chordoma = CS | 2.0 | NA** | 0.14 | |
| CD24 | CD24 antigen | Chordoma > CS | 36.3 | 27.3 | 0.01 |
| CD24 | CD24 antigen | Chordoma > STS | 50.6 | 26.5 | 0.009 |
| T | T, Brachyury | Chordoma > CS | 1072.9 | 228.1 | 0.03 |
| T | T, Brachyury | Chordoma > STS | 17157.3 | 79.7 | 0.03 |
P values are based on RT-PCR data for the respective groups compared;
NA HMW-MAA gene was not differentially expressed in chordoma and chondrosarcoma samples by microarray analysis
HMW-MAA protein expression in more than half of the chordoma and chondrosarcoma tumors tested, as well as in human chondrosarcoma and chordoma cell lines
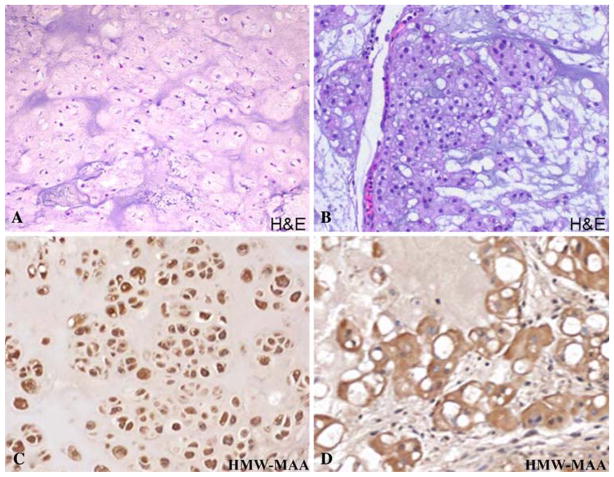
Immunohistochemical staining with HMW-MAA-specific mAb resulted in the staining of 62 and 48% of the 21 chordoma and 84 chondrosarcoma lesions tested, respectively. The staining correlated neither with the histologic grade nor dedifferentiation of the chordoma and chondrosarcoma lesions tested. The staining pattern observed was a diffuuse and strong cytoplasmic reactivity in the majority of the tumor cells of the positive cases of both chordoma and chondrosarcoma samples (Fig. 3).
Fig. 3.
a Hematoxylin and eosin staining of a chondrosarcoma lesion demonstrating hyaline matrix (H&E, ×200); b Hematoxylin and eosin staining of a chordoma lesion with characteristic mucinous matrix and physaliferous cells (H&E, ×200); c Immunohistochemical staining of a chondrosarcoma sample with HMW-MAA specific mAb demonstrating strong and diffuuse cytoplasmic positivity (×200). d Immunohistochemical staining of a chordoma with HMW-MAA specific mAb demonstrating strong cytoplasmic reactivity (×200)
Quantitative real-time PCR found that the HMW-MAA mRNA level was increased in both chordoma and chondrosarcoma samples relative to soft tissue sarcoma lesions, but was not significantly different between chordoma and chondrosarcoma tumors (P = 0.14) (Table 2).
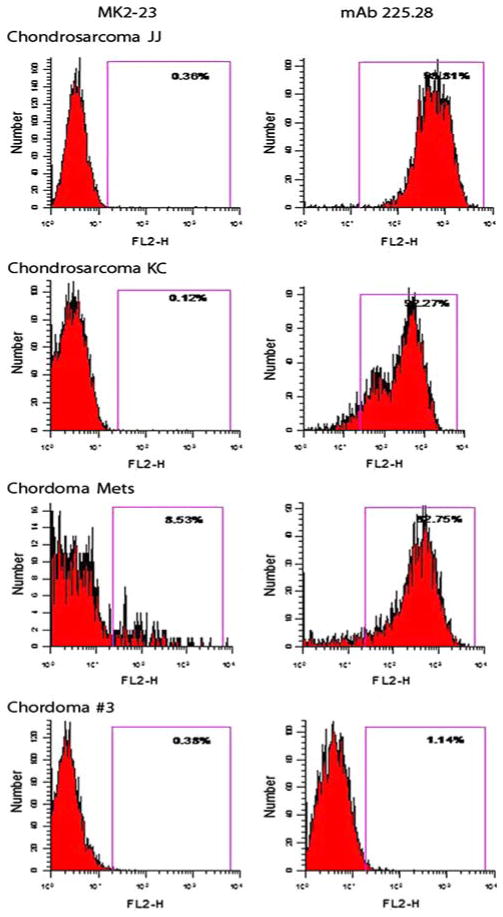
FACS analysis of human JJ and KC human chondrosarcoma cell lines and of the human chordoma metastatic cell line stained with HMW-MAA-specific mAb 116, 149.53, 225.28, 724, 763.74, TP32, TP41.2, TP43, TP61.5, TP108, TP109, VF18.176 VT1.7, VT5-1, VT67.5, VT68.2 and VT80.12 showed that the three cell lines were stained by all the HMW-MAA-specific mAbs tested. These results indicate that, at variance with the results obtained with melanoma cells lines [9], all the determinants recognized by the mAb tested are expressed on the HMW-MAA synthesized by the chordoma and chondrosarcoma cell lines analyzed. However, the human primary chordoma cell line Chordoma #3 was not stained by any of the HMW-MAA-specific mAb tested. Representative results are shown in Fig. 4.
Fig. 4.
Flow cytometric analysis of chondrosarcoma and chordoma cell lines stained with the HMW-MAA-specific mAb 225.28. The irrelevant mouse anti-idiotypic mAb MK2-23 was used as a specificity control
Western blotting analysis with HMW-MAA-specific mAb 763.74 showed that the molecular profile of the HMW-MAA synthesized by chondrosarcoma cell lines and by the metastatic chordoma cell line tested was similar to that of the antigen synthesized by the cultured human melanoma cell line. On the other hand, no component was detected in the lysate of the Chordoma cell line #3.
Discussion
Both chordomas and chondrosarcomas are notoriously resistant to conventional therapy modalities. This drug resistance might be secondary to the abundant matrix protecting the individual tumor cells from systemic delivery of chemotherapy. Conversely, the malignant cells must overcome the matrix barrier to metastasize. The extracellular matrix contains highly negative-charged glycosaminoglycans, including aggrecan and chondroitin, heparin and dermatan-type sulfates. It is the interface between the matrix and the individual cells that may prove to be an important target for systemic therapy. Cleavage of the matrix proteins requires enzymatic degradation and several metalloproteinase genes are overexpressed in both tumor types, including MMP-9, 19, ADAM28 and ADAMTS9. ADAMTS9 is an aggrecanase which is responsive to IL-1 and TNF-α stimulation in chondrosarcoma cell lines [11]. MMP-9 (gelatinase B) degrades denatured collagen and basement membrane proteins [33] and is known to be expressed in chordomas [26]. In addition, MMP-3, 7 and 13 are highly expressed in chondrosarcoma samples. MMP-13 has been previously shown to be present in chondrosarcoma cells in vitro [32, 42]. Targeted molecular therapies against metalloproteinases have recently become an area of intense research [28]. Inhibition of these proteases could prevent the local growth and distant spread of these tumors, however, matrix degradation is only one requisite needed for tumor metastasis. One cell surface proteoglycan known to be involved in cell–matrix interactions is HMW-MAA, the function of which is linked to cellular migration and invasion; both of them are necessary for tumor cells to grow and spread systemically [22]. Our present study showed HMW-MAA expression in a significant number of both chondrosarcoma and chordoma tumors, as compared to normal tissues and in a wide variety of other histologic types of sarcomas.
Although all chordoma samples included in our study had a conventional morphology and lacked chondroid differentiation, they consistently expressed genes commonly found in cartilage, including type II collagen, aggrecan, SOX 9, and CD-RAP. SOX 9 is a critical transcription factor essential for chondrogenesis, and known to be involved in the transcription of type II collagen, aggrecan and CD-RAP [5, 15, 21, 38, 43, 45]. Matrillin 3, expressed in both chordoma and chondrosarcoma lesions, encodes a non-collagenous, extracellular matrix protein found in normal cartilage and serves as a link between collagen containing fibrils and glycosaminoglycans, such as aggrecan [44].
Components of the activator protein 1 (AP-1) transcription factor were highly expressed in both chordoma and chondrosarcoma relative to various soft tissue sarcomas and also to nucleus pulposus and articular cartilage tissue. The same pattern of expression was seen with FOSB, c-Fos and c-Jun. Deregulation of AP-1 transcription factor is thought to be sufficient for tumorigenesis, and AP-1 is considered critical in the function of dominant oncogenes [30]. Several genes known to be regulated by AP-1 were highly expressed in our samples, including EGFR, MMP3 and MMP9. [8, 36].
Chordoma-specific genes included T Brachyury, CD24, Keratin 8, 13, 15, 18, 19 and discoidin domain receptor 1. This expression profile is in concordance with the findings obtained by Vujovic et al. [43] who compared a group of chordoma lesions to a variety of chondroid tumors, including chondrosarcomas, although the specific breakdown of each type of tumor, in terms of number of cases and histologic subtype, was not specified. Our study found these genes highly expressed relative to a variety of soft tissue sarcomas as well as to nucleus pulposus tissue. The significantly higher expression of T Brachyury in chordoma versus nucleus pulposus (FC, 38) is in keeping with previous immunohistochemical studies that did not identify T Brachyury in nucleus pulposus.
Chondrosarcomas were distinguished relative to benign articular cartilage, chordomas, and soft tissue sarcomas by their expression of Type IX and XI collagen genes. Both encode fibrillar collagens expressed during cartilage development and form important links to type II collagen [12]. Up-regulation of several additional non-collagenous matrix genes, such as Cartilage Oligomeric Matrix Protein (COMP) and Matrillin 1, stood out in our analysis of chondrosarcoma lesions. In contrast with the results reported by Vujovic et al. [43], we did not find Platelet Derived Growth Factor A or type X collagen to be preferentially overexpressed in chondrosarcomas. Type X collagen, thought to be involved in cartilage calcification, was not detected in the chordoma samples by prior investigators and was correlated with the lack of calcification noted in these tumors [15, 43]. However, Type X collagen and bone sialoprotein are normally found together in the hypertrophic zone of the physis [10], and their genes were up-regulated at high levels in both chordoma and chondrosarcoma lesions in our study. While type X collagen is associated with chondroid calcification, its presence or absence has not been proven to be required for cartilage calcification. These discrepancies are likely due to differences in study design and control groups used for analysis.
In conclusion, chordoma and chondrosarcoma express a similar subset of genes involved in extracellular matrix synthesis and control, which overshadow the limited number of up-regulated tumor-type specific genes. These findings suggest that targeting cell–matrix interaction might be a promising therapeutic strategy for these tumors. Preventing the egress of cells from the extracellular matrix by inhibiting proteases or blocking proteins important for cell mobilization may be a tenable alternative approach to conventional systemic treatment modalities. In this regard, a potential target is HMW-MAA, which is highly expressed in both chordoma and chondrosarcoma lesions. Targeting HMW-MAA interaction with extracellular matrix proteins might prove beneficial in these chemo-refractory malignant bone tumors, by decreasing cell migration through their abundant extracellular matrix. This possibility is supported by the association we have found between the development of HMW-MAA-specific antibodies in patients with melanoma immunized with a HMW-MAA mimic and regression of metastases in a few patients and a statistically significant survival prolongation [24, 25].
Contributor Information
Joseph H. Schwab, Department of Surgery, Orthopedic Service, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Patrick J. Boland, Department of Surgery, Orthopedic Service, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Narasimhan P. Agaram, Department of Pathology, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10021, USA
Nicholas D. Socci, Computational Biology Center, Sloan-Kettering Institute, New York, NY, USA
Tianhua Guo, Department of Pathology, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10021, USA.
Gary C. O’Toole, Department of Surgery, Orthopedic Service, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Xinhui Wang, Department of Immunology, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA.
Elena Ostroumov, Department of Mechanical and Manufacturing Engineering Centre for Bioengineering Research and Education, University of Calgary, Calgary, AB, Canada.
Christopher J. Hunter, Department of Mechanical and Manufacturing Engineering Centre for Bioengineering Research and Education, University of Calgary, Calgary, AB, Canada
Joel A. Block, Rheumatology Section, Rush University Medical Center, Chicago, IL, USA
Stephen Doty, The Hospital for Special Surgery, New York, NY, USA.
Soldano Ferrone, Departments of Surgery, of Immunology and of Pathology, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA.
John H. Healey, Department of Surgery, Orthopedic Service, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Cristina R. Antonescu, Email: antonesc@mskcc.org, Department of Pathology, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10021, USA
References
- 1.Antonescu CR, Viale A, Sarran L, Tschernyavsky SJ, Gonen M, Segal NH, Maki RG, Socci ND, DeMatteo RP, Besmer P. Gene expression in gastrointestinal stromal tumors is distinguished by KIT genotype and anatomic site. Clin Cancer Res. 2004;10:3282–3290. doi: 10.1158/1078-0432.CCR-03-0715. [DOI] [PubMed] [Google Scholar]
- 2.Agaram NP, Besmer P, Wong GC, Guo T, Socci ND, Maki RG, DeSantis D, Brennan MF, Singer S, DeMatteo RP, Antonescu CR. Pathologic and molecular heterogeneity in imatinib-stable or imatinib-responsive gastrointestinal stromal tumors. Clin Cancer Res. 2007;13:170–181. doi: 10.1158/1078-0432.CCR-06-1508. [DOI] [PubMed] [Google Scholar]
- 3.Bergh P, Kindblom LG, Gunterberg B, Remotti F, Ryd W, Meis-Kindblom JM. Prognostic factors in chordoma of the sacrum and mobile spine: a study of 39 patients. Cancer. 2000;88:2122–2134. doi: 10.1002/(sici)1097-0142(20000501)88:9<2122::aid-cncr19>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Bertoni F, Bacchini P, Hogendoorn P. Chondrosarcoma. IARC Press; Lyon: 2002. [Google Scholar]
- 5.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 6.Bjornsson J, McLeod RA, Unni KK, Ilstrup DM, Pritchard DJ. Primary chondrosarcoma of long bones and limb girdles. Cancer. 1998;83:2105–2119. [PubMed] [Google Scholar]
- 7.Block JA, Inerot SE, Gitelis S, Kimura JH. The effects of long term monolayer culture on the proteoglycan phenotype of a clonal population of mature human malignant chondrocytes. Connect Tissue Res. 1991;26:295–313. doi: 10.3109/03008209109152446. [DOI] [PubMed] [Google Scholar]
- 8.Bos TJ, Margiotta P, Bush L, Wasilenko W. Enhanced cell motility and invasion of chicken embryo fibroblasts in response to Jun over-expression. Int J Cancer. 1999;81:404–410. doi: 10.1002/(sici)1097-0215(19990505)81:3<404::aid-ijc14>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 9.Campoli MR, Chang CC, Kageshita T, Wang X, McCarthy JB, Ferrone S. Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit Rev Immunol. 2004;24:267–296. doi: 10.1615/critrevimmunol.v24.i4.40. [DOI] [PubMed] [Google Scholar]
- 10.de Bri E, Reinholt FP, Heinegard D, Mengarelli-Widholm S, Norgard M, Svensson O. Bone sialoprotein and osteopontin distribution at the osteocartilaginous interface. Clin Orthop Relat Res. 1996;330:251–260. doi: 10.1097/00003086-199609000-00033. [DOI] [PubMed] [Google Scholar]
- 11.Demircan K, Hirohata S, Nishida K, Hatipoglu OF, Oohashi T, Yonezawa T, Apte SS, Ninomiya Y. ADAMTS-9 is synergistically induced by interleukin-1beta and tumor necrosis factor alpha in OUMS-27 chondrosarcoma cells and in human chondrocytes. Arthritis Rheum. 2005;52:1451–1460. doi: 10.1002/art.21010. [DOI] [PubMed] [Google Scholar]
- 12.Eyre DR, Wu JJ, Fernandes RJ, Pietka TA, Weis MA. Recent developments in cartilage research: matrix biology of the collagen II/IX/XI heterofibril network. Biochem Soc Trans. 2002;30:893–899. doi: 10.1042/bst0300893. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs B, Dickey ID, Yaszemski MJ, Inwards CY, Sim FH. Operative management of sacral chordoma. J Bone Joint Surg Am. 2005;87:2211–2216. doi: 10.2106/JBJS.D.02693. [DOI] [PubMed] [Google Scholar]
- 14.Gitelis S, Bertoni F, Picci P, Campanacci M. Chondrosarcoma of bone. The experience at the Istituto Ortopedico Rizzoli. J Bone Joint Surg Am. 1981;63:1248–1257. [PubMed] [Google Scholar]
- 15.Gottschalk D, Fehn M, Patt S, Saeger W, Kirchner T, Aigner T. Matrix gene expression analysis and cellular phenotyping in chordoma reveals focal differentiation pattern of neoplastic cells mimicking nucleus pulposus development. Am J Pathol. 2001;158:1571–1578. doi: 10.1016/S0002-9440(10)64111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Healey JH, Lane JM. Chondrosarcoma. Clin Orthop Relat Res. 1986:119–129. [PubMed] [Google Scholar]
- 17.Healey JH, Lane JM. Chordoma: a critical review of diagnosis and treatment. Orthop Clin North Am. 1989;20:417–426. [PubMed] [Google Scholar]
- 18.Henderson SR, Guiliano D, Presneau N, McLean S, Frow R, Vujovic S, Anderson J, Sebire N, Whelan J, Athanasou N, Flanagan AM, Boshoff C. A molecular map of mesenchymal tumors. Genome Biol. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida T, Dorfman HD. Chondroid chordoma versus low-grade chondrosarcoma of the base of the skull: can immunohistochemistry resolve the controversy? J Neurooncol. 1994;18:199–206. doi: 10.1007/BF01328954. [DOI] [PubMed] [Google Scholar]
- 20.Kusama M, Kageshita T, Chen ZJ, Ferrone S. Characterization of syngeneic antiidiotypic monoclonal antibodies to murine anti-human high molecular weight melanoma-associated antigen monoclonal antibodies. J Immunol. 1989;143:3844–3852. [PubMed] [Google Scholar]
- 21.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo W, Ko E, Hsu JC, Wang X, Ferrone S. Targeting melanoma cells with human high molecular weight-melanoma associated antigen-specific antibodies elicited by a peptide mimotope: functional effects. J Immunol. 2006;176:6046–6054. doi: 10.4049/jimmunol.176.10.6046. [DOI] [PubMed] [Google Scholar]
- 23.Mirra J, Rocca C, Nelson S, Mertens F. Notochordal tumours. IARC Press; Lyon: 2002. [Google Scholar]
- 24.Mittelman A, Chen ZJ, Yang H, Wong GY, Ferrone S. Human high molecular weight melanoma-associated antigen (HMW-MAA) mimicry by mouse anti-idiotypic monoclonal antibody MK2-23: induction of humoral anti-HMW-MAA immunity and prolongation of survival in patients with stage IV melanoma. Proc Natl Acad Sci USA. 1992;89:466–470. doi: 10.1073/pnas.89.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittelman A, Chen ZJ, Liu CC, Hirai S, Ferrone S. Kinetics of the immune response and regression of metastatic lesions following development of humoral anti-high molecular weight-melanoma associated antigen immunity in three patients with advanced malignant melanoma immunized with mouse antiidiotypic monoclonal antibody MK2-23. Cancer Res. 1994;54:415–421. [PubMed] [Google Scholar]
- 26.Naka T, Boltze C, Kuester D, Schulz TO, Samii A, Herold C, Ostertag H, Roessner A. Expression of matrix metalloproteinase (MMP)-1, MMP-2, MMP-9, cathepsin B, and urokinase plasminogen activator in non-skull base chordoma. Am J Clin Pathol. 2004;122:926–930. doi: 10.1309/C8T7-APJD-AUPR-8TLL. [DOI] [PubMed] [Google Scholar]
- 27.Ostroumov E, Hunter CJ. The role of extracellular factors in human metastatic chordoma cell growth in vitro. Spine. 2007;32:2957–2964. doi: 10.1097/BRS.0b013e31815cde91. [DOI] [PubMed] [Google Scholar]
- 28.Overall CM, Kleifeld O. Tumour microenvironment—opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 29.Ozaki T, Hillmann A, Winkelmann W. Surgical treatment of sacrococcygeal chordoma. J Surg Oncol. 1997;64:274–279. doi: 10.1002/(sici)1096-9098(199704)64:4<274::aid-jso5>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 31.Palena C, Polev DE, Tsang KY, Fernando RI, Litzinger M, Krukovskaya LL, Baranova AV, Kozlov AP, Schlom J. The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell-mediated cancer immunotherapy. Clin Cancer Res. 2007;13:2471–2478. doi: 10.1158/1078-0432.CCR-06-2353. [DOI] [PubMed] [Google Scholar]
- 32.Pei Y, Harvey A, Yu XP, Chandrasekhar S, Thirunavukkarasu K. Differential regulation of cytokine-induced MMP-1 and MMP-13 expression by p38 kinase inhibitors in human chondrosarcoma cells: potential role of Runx2 in mediating p38 effects. Osteoarthritis Cartilage. 2006;14:749–758. doi: 10.1016/j.joca.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Polette M, Nawrocki-Raby B, Gilles C, Clavel C, Birembaut P. Tumour invasion and matrix metalloproteinases. Crit Rev Oncol Hematol. 2004;49:179–186. doi: 10.1016/j.critrevonc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Pring ME, Weber KL, Unni KK, Sim FH. Chondrosarcoma of the pelvis. A review of sixty-four cases. J Bone Joint Surg Am. 2001;83-A:1630–1642. [PubMed] [Google Scholar]
- 35.Rosenberg AE, Brown GA, Bhan AK, Lee JM. Chondroid chordoma—a variant of chordoma. A morphologic and immunohistochemical study. Am J Clin Pathol. 1994;101:36–41. doi: 10.1093/ajcp/101.1.36. [DOI] [PubMed] [Google Scholar]
- 36.Scott LA, Vass JK, Parkinson EK, Gillespie DA, Winnie JN, Ozanne BW. Invasion of normal human fibroblasts induced by v-Fos is independent of proliferation, immortalization, and the tumor suppressors p16INK4a and p53. Mol Cell Biol. 2004;24:1540–1559. doi: 10.1128/MCB.24.4.1540-1559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal NH, Pavlidis P, Antonescu CR, Maki RG, Noble WS, DeSantis D, Woodruff JM, Lewis JJ, Brennan MF, Houghton AN, Cordon-Cardo C. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 39.Sundaresan N, Galicich JH, Chu FC, Huvos AG. Spinal chordomas. J Neurosurg. 1979;50:312–319. doi: 10.3171/jns.1979.50.3.0312. [DOI] [PubMed] [Google Scholar]
- 40.Temponi M, Kageshita T, Perosa F, Ono R, Okada H, Ferrone S. Purification of murine IgG monoclonal antibodies by precipitation with caprylic acid: comparison with other methods of purification. Hybridoma. 1989;8:85–95. doi: 10.1089/hyb.1989.8.85. [DOI] [PubMed] [Google Scholar]
- 41.Tirabosco R, Mangham DC, Rosenberg AE, Vujovic S, Bousdras K, Pizzolitto S, De Maglio G, den Bakker MA, Di Francesco L, Kalil RK, Athanasou NA, O’Donnell P, McCarthy EF, Flanagan AM. Brachyury expression in extra-axial skeletal and soft tissue chordomas: a marker that distinguishes chordoma from mixed tumor/myoepithelioma/parachordoma in soft tissue. Am J Surg Pathol. 2008;32(4):572–580. doi: 10.1097/PAS.0b013e31815b693a. [DOI] [PubMed] [Google Scholar]
- 42.Uria JA, Balbin M, Lopez JM, Alvarez J, Vizoso F, Takigawa M, Lopez-Otin C. Collagenase-3 (MMP-13) expression in chondrosarcoma cells and its regulation by basic fibroblast growth factor. Am J Pathol. 1998;153:91–101. doi: 10.1016/S0002-9440(10)65549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vujovic S, Henderson S, Presneau N, Odell E, Jacques TS, Tirabosco R, Boshoff C, Flanagan AM. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209:157–165. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- 44.Wagener R, Ehlen HW, Ko YP, Kobbe B, Mann HH, Sengle G, Paulsson M. The matrilins-adaptor proteins in the extracellular matrix. FEBS Lett. 2005;579:3323–3329. doi: 10.1016/j.febslet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 46.Zharkikh A, Li WH. Estimation of confidence in phylogeny: the complete-and-partial bootstrap technique. Mol Phylogenet Evol. 1995;4:44–63. doi: 10.1006/mpev.1995.1005. [DOI] [PubMed] [Google Scholar]