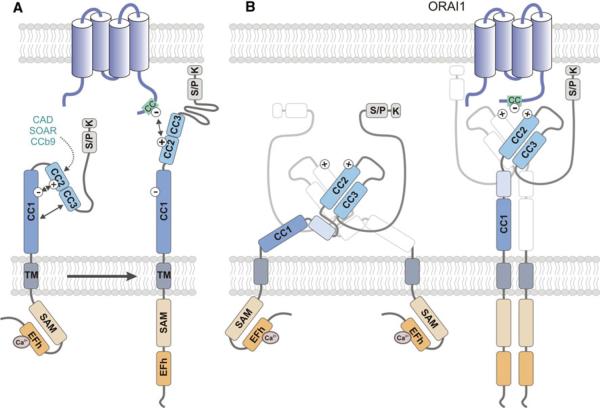
Fig. 3.
Conformational changes of STIM1 structure during CRAC channel activation. In non-stimulated cells, ER Ca2+ stores are filled and Ca2+ is bound to the cEFh domain in the NT of STIM1. Upon depletion of Ca2+ from ER stores, the [Ca2+]ER drops and Ca2+ dissociates from the EFh, resulting in a conformational change of the STIM1-NT. Structural data support a model in which the Ca2+-free EFh unfolds from the adjacent SAM domain [42], thereby allowing the interaction of SAM domains in neighboring STIM1 molecules and their oligomerization. The STIM1-CT and its CC domains are critical for STIM1 oligomerization and ORAI1 binding. The structure of the STIM1-CT and the conformational changes that occur within it following store depletion are incompletely defined. a In one model, the STIM1-CT is present in a folded, inactive conformation in cells with replete ER stores. CC2 and CC3 (which encompass the majority of the CAD/CCb9/SOAR domain) were proposed to interact with CC1 (which was shown to inhibit the ability of the STIM1-CT to activate CRAC channels) through hydrophobic interactions and hydrogen bonds between acidic (CC1: 302–322) and basic (CC2: 382–386) residues (indicated by minus and plus symbols) [58, 63, 64, 66]. Upon store depletion, the STIM1-CT unfolds, allowing STIM1 oligomerization. b An alternative model proposed by Yang et al. [68] is based on the crystal structure of the human CAD domain and the CC1-CAD domains of C. elegans. The CAD domain, which contains two long alpha helices aligned in an antiparallel manner, exists as a dimer in solution. Dimerization is stabilized by hydrophobic interactions, hydrogen bonds, and stacking interactions between residues located at the N- and C-terminal ends of each CAD domain (for details see text). The CAD dimer forms a V-shaped structure that exposes basic Lys and Arg residues (aa 382–387; indicated by plus symbols), which may mediate binding to ORAI1 as reported earlier [63, 66]. CAD function is restrained by an inhibitory helix at the C-terminal end of CC1 (light blue box) that interacts with CAD in several locations [68]. STIM1 activation is mediated by release of the inhibitory helix from CAD. In contrast to the model shown in a, the structure of the CAD dimer remains unchanged after store depletion. For clarity, only one of the dimer-forming CAD domains is shown in color. CC coiled-coil, CT C-terminus, EFh EF-hand, K lysine-rich, NT N terminus, SAM sterile alpha motif, S/P serine/proline-rich, TM transmembrane