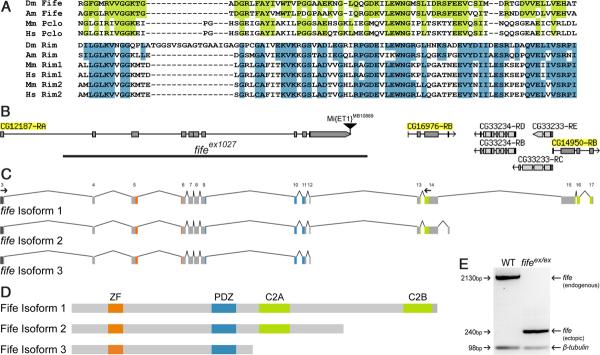
Figure 1.
Molecular identification of a piccolo-rim-related gene in Drosophila. A, PDZ domain of Fife/Piccolo (top) and RIM (bottom) family proteins are aligned to illustrate conservation among the families. Highlighted regions indicate identity between the Drosophila protein and vertebrate Piccolo (green) or RIM (blue). (Am), A. mellifera; (Dm), Drosophila melanogaster; (Hs), Homo sapiens; (Mm), Mus musculus. B, Full-length fife comprises 17 exons corresponding to a predicted open reading frame previously annotated as three separate candidate genes CG12187, CG16976, and CG14950. Two 5′ UTR exons, located 10 and 20 kb upstream, are not depicted. fifeex1027 was generated by excision of the Minos element Mi{ET1}CG43375MB10889. The location of the 16.5 deletion is indicated (black bar). C, fife encodes three transcripts. Exons are numbered above isoform 1. D, Full-length Fife (Isoform 1) has four conserved functional domains: ZF (orange), PDZ (blue), C2A (green), and C2B (green). Alternative isoforms lack either the C2B domain (Isoform 2) or both C2 domains (Isoform 3). E, Real-time PCR products from amplification of exons 3–14 in wild type (WT) and fifeex1027. Primer locations are indicated by small arrows in C. A full-length fife product of 2130 bp is amplified from wild-type, but not fifeex1027, cDNA. Conversely, a 240 bp band representing an aberrant splicing event between exons 3 and 13 is present in fifeex1027 but not in wild type. β-tubulin was also amplified in these reactions as a loading control. B is modified from FlyBase (McQuilton et al., 2012).