Fig. 6.
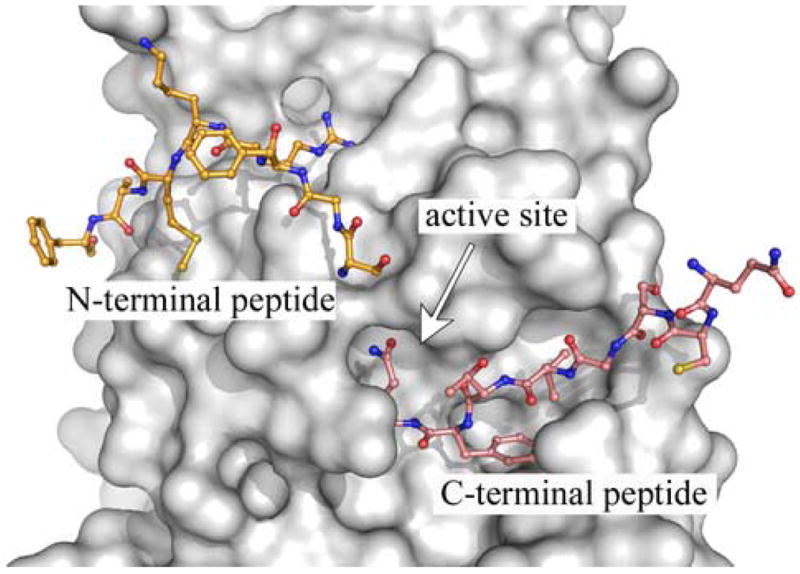
X-ray structure of a substrate-like complex of SARS-CoV 3CLpro. A single monomer of a Cys145Ala mutant of the SARS-CoV 3CLpro dimer, having extended amino acid sequences at the n- and c-terminus, is shown as a surface representation (grey). The uncleaved, n-terminus of the second monomer of the dimer (yellow) extends into the active site. In addition, the c-terminus from a monomer in another asymmetric (cyan) also extends into the active site. The figure was made from PDB accession number 1Z1J (Hsu et al., 2005), using the program PyMOL version 0.99 (DeLano Scientific, LLC).
