Abstract
While Rhodopsin is essential for sensing light for vision, it also mediates light-induced apoptosis of photoreceptors in mouse. RPE65, which catalyzes isomerization of all-trans retinyl fatty acid esters to 11-cis retinol (11cROL) in the visual cycle, controls the rhodopsin regeneration rate and photoreceptor susceptibility to light-induced degeneration. Mutations in RPE65 have been linked to blindness in affected children. Despite such importance, the mechanism that regulates RPE65 function remains unclear. Through unbiased expression screening of a bovine retinal pigment epithelium (RPE) cDNA library, we have identified elongation of very long-chain fatty acids-like 1 (ELOVL1) and fatty acid transport protein 4 (FATP4), which each have very long-chain fatty acid acyl-CoA synthetase (VLCFA-ACS) activity, as negative regulators of RPE65. We found that the VLCFA derivative lignoceroyl (C24:0)-CoA inhibited synthesis of 11cROL, whereas palmitoyl (C16:0)-CoA promoted synthesis of 11cROL. We further found that competition of FATP4 with RPE65 for the substrate of RPE65 was also involved in the mechanisms by which FATP4 inhibits synthesis of 11cROL. FATP4 was predominantly expressed in RPE, and the FATP4-deficient RPE showed significantly higher isomerase activity. Consistent with these results, the regeneration rate of 11-cis retinaldehyde and the recovery rate for rod light sensitivity were faster in FATP4-deficient mice than wild-type mice. Moreover, FATP4-deficient mice displayed increased accumulation of the cytotoxic all-trans retinaldehyde and hyper susceptibility to light-induced photoreceptor degeneration. Our findings demonstrate that ELOVL1, FATP4, and their products comprise the regulatory elements of RPE65 and play important roles in protecting photoreceptors from degeneration induced by light damage.
Introduction
The visual cycle provides 11-cis retinaldehyde (11cRAL) chromophore to photoreceptors to regenerate visual pigments that sense light. Although the visual cycle is essential for sustaining vision, its all-trans retinaldehyde (atRAL) intermediate possesses a highly reactive aldehyde group and is toxic to photoreceptors (Maeda et al., 2009). Mice with a slow visual cycle contain lower amounts of cytotoxic byproducts of the visual cycle (Kim et al., 2004). Pharmacological inhibition of the visual cycle protects photoreceptors from degeneration in physiological and pathological conditions (Sieving et al., 2001; Golczak et al., 2008), suggesting that the visual cycle needs to be controlled to provide enough 11cRAL chromophore and to keep retinal health. However, the mechanisms that regulate the cycle remain unclear. Identification and characterization of regulator(s) of the visual cycle enzymes are critical for understanding these mechanisms.
RPE65 is a key enzyme that isomerizes all-trans retinyl esters (atRE), such as all-trans retinyl palmitate (atRP), to 11-cis retinol (11cROL) (Jin et al., 2005; Moiseyev et al., 2005; Redmond et al., 2005), which constitutes the rate-limiting step of the visual cycle (Winston and Rando, 1998). The palmitic acyl moiety of atRP is essential for its binding to RPE65 (Maiti et al., 2005) and may facilitate substrate access to the catalytic site located inside a hydrophobic pocket of RPE65 (Kiser et al., 2009). These facts suggest that protein(s) interacting with the fatty acyl moiety regulate RPE65 function. A palmitoylation mechanism for regulation of isomerase activity has been proposed (Xue et al., 2004), but this mechanism is not consistent with results reported by later studies (Redmond et al., 2005; Jin et al., 2007; Kiser et al., 2009; Yuan et al., 2010).
Mutations in RPE65 are associated with blinding diseases (Gu et al., 1997; Marlhens et al., 1997). Disease-causing mutations severely reduce isomerase activity (Chen et al., 2006; Takahashi et al., 2006; Philp et al., 2009). Mice with a null mutation in Rpe65 cannot synthesize 11cROL and thereby lack functional rhodopsin (Redmond et al., 1998; Pang et al., 2005). Rpe65+/− mice show slowing of 11cRAL and rhodopsin regeneration. Either the lack or slowed regeneration of 11cRAL and rhodopsin protected photoreceptors from apoptosis induced by light (Grimm et al., 2000; Wenzel et al., 2001). In contrast, mice with higher expression levels of RPE65 exhibited hyper-susceptibility to light-induced retinal degeneration (Wenzel et al., 2001). RPE65 is highly abundant in the retinal pigment epithelium (RPE), yet its activity is significantly lower than that of other visual cycle enzymes. For instance, the Vmax for the lecithin:retinol acyltransferase (LRAT) in bovine RPE was 103–199 nmol/min/mg (Saari and Bredberg, 1988), whereas the Vmax for the isomerase was 44.3 pmol/min/mg (Winston and Rando, 1998). The mechanisms responsible for this low activity are poorly understood. One possible hypothesis is the existence of inhibitor(s) of RPE65 in RPE. Here, we identified elongation of very long-chain fatty acids-like 1 (ELOVL1) and fatty acid transport protein 4 (FATP4) as inhibitors of RPE65. We further analyzed the inhibitory mechanisms of FATP4 and the phenotypes of mice lacking FATP4 in RPE.
Materials and Methods
Animals
129S2/Sv (Charles River Laboratories), C57BL/6J (The Jackson Laboratory) and Fatp4−/−;Ivl-Fatp4tg/+ (Moulson et al., 2007) mice were maintained in 12 h cyclic light at 30 lux. Fatp4−/−;Ivl-Fatp4tg/+ mice (shown as Fatp4−/− hereafter) are transgenic/mutant mice expressing transgenic FATP4 in keratinocytes (via the involucrin promoter) of a FATP4-deficient mouse line called wrinkle free, which has a spontaneous retrotransposon insertion in a coding exon of Fatp4 (Moulson et al., 2003). The skin defect-based neonatal lethality of Fatp4−/− wrinkle free mice is rescued by keratinocyte-specific expression of FATP4 (Moulson et al., 2007). Because the original Fatp4−/− mice were heterozygous for the Leu450Met variation in Rpe65, we crossed them with 129S2/Sv and C57BL/6J mice, then intercrossed the heterozygous offspring to yield Fatp4−/− mice homozygous for the Leu450 or Met450 allele. The Leu450 and Met450 alleles were verified by DNA sequencing using Rpe65-specific primers (5’-CAGAAATTTGGAGGGAAACC-3’ and 5’-TACCATCATCTTCTTCCAGAGC-3’). The Fatp4−/− mutation was confirmed by PCR as described previously (Moulson et al., 2007). 129S2/Sv and C57BL/6J mice were used as wild-type (WT) controls against age-matched Fatp4−/− mice with Leu450 or Met450 alleles, respectively. Two to three month old mice of either sex were used for the experiments unless otherwise specified. All mouse experiments were approved by the Institutional Animal Care and Use Committee for LSUHSC and performed according to guidelines established by the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Cell culture and transfection
The 293T-LRC cells (Jin et al., 2005) stably expressing LRAT (L), RPE65 (R), and the cellular retinaldehyde-binding protein (C) were maintained in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (100 units/ml penicillin G and 100 µg/ml streptomycin) at 37°C under 5% CO2. Transient transfection of plasmid DNA into the cells was performed using the PolyJet transfection reagent (SignaGen Laboratories).
Screening of the RPE expression library
Library screening was done as described previously (Jin et al., 2005). The library pool #7:3 that produced the lowest amount of 11cROL in the transfected cells (Jin et al., 2005) was split onto 32 dishes at an average density of 39 colonies per dish for tertiary screening. Plasmid DNA was prepared using the PureLink HiPure Plasmid Kit (Invitrogen). Two micrograms of library DNA from each dish and 100 ng of pRPE65 were cotransfected into 293T-LRC cells grown in 6-well dishes for the retinoid isomerase assay. Cells transfected with 2 µg of pRK5 mock vector and 100 ng of pRPE65 were used as a control. Cotransfection of pRPE65 increased baseline of the isomerase activity in the cells and thereby facilitated selection of a library pool with the strongest inhibitory effect on the isomerase activity. For the final round of screening, 0.5 µg of individual clones and the same amount of pRPE65 were cotransfected into the cells grown in 12-well dishes. Thirty hours after transfection, cells were incubated with 5 µM atROL for 16 h. Retinoids extracted from the cells were saponified and analyzed by HPLC as described previously (Jin et al., 2005).
Expression of recombinant proteins in the Sf9 cells
Baculovirus-mediated expression of recombinant RPE65, FATP4, and EGFP in Sf9 cells was performed as previously described (Jin et al., 2000). Briefly, cDNAs for RPE65, FATP4, and EGFP were subcloned into pBAC-1 (Novagen). These transfer vectors were then cotransfected into Sf9 cells with a triple-cut baculovirus DNA (Novagen) using Cellfectin (Invitrogen). The recombinant baculovirus clones expressing FATP4, RPE65, or EGFP were isolated by plaque purification. High-titer (109 plaque-forming units per ml, PFU/ml) virus stocks were generated in the Sf9 cells. Three days after infection with the recombinant viruses at the desired multiplicity of infection (MOI), the cells were harvested and used for enzyme assays. Expression levels of recombinant proteins were determined by immunoblot analysis.
In vitro retinoid isomerase assay
The assay mixtures contained 10 µM atROL (or 15 µM all-trans retinyl palmitate), 6% bovine serum albumin, and 100 µg of mouse RPE homogenates or 500 µg of Sf9 cell homogenates in 20 mM HEPES buffer. When atRP was used as a substrate, 6 mM sodium cholate was added to the reaction mixture. For enzyme kinetic assays, Sf9 cells containing a constant amount of RPE65 plus various amounts of FATP4 or EGFP were incubated with 0.5–64 µM of atRP for 90 min. Retinoids were extracted with hexane and were analyzed by HPLC, as described below. The quantified data were then compared in the different inhibition models using the Enzyme Kinetics Module 1.3.
LRAT and retinol dehydrogenase 5 (RDH5) assays in living cells
293T-LRC cells grown in a 12-well dish were cotransfected with pRPE65 plus either FATP4, ELOVL1, RDH5, or pRK5 (mock vector), and were incubated with 10 µM of atROL for 0.5–4 h. To determine the activity of LRAT, the amount of atRE synthesized in the cells was measured as described previously (Jin et al., 2007). The activity of RDH5 was determined by measuring the synthesis of 11cRAL in the cells. We converted 11cRAL to syn- and anti-retinaloxime isomers using 150 mM hydroxylamine prior to extraction of retinoids from the cells. Measurement of 11cRAL was done as described previously (Jin et al., 2009).
Analysis of retinoids
Retinoids were analyzed by normal-phase high-performance liquid chromatography (HPLC) as previously described (Mata et al., 2002). In brief, retinoids in hexane extractions were evaporated, re-dissolved in 100 µl of hexane, and separated on a silica column (Zorbax-Sil 5 µm, 250×4.6 mm, Agilent Technologies) by gradient (0.2–10% dioxane in hexane at 2.0 ml/min flow rate) or non-gradient (10% dioxane in hexane at 1.0 ml/min flow rate) elution of mobile phase in an Agilent 1100 HPLC system.
Analysis of 11cRAL and atRAL contents in the retina
Overnight dark-adapted mice were anesthetized with an intraperitoneal injection of ketamine-xylazine mixture (200 mg ketamine and 10 mg xylazine per kilogram body weight) and their pupils dilated with 1% tropicamide ophthalmic solution. Mice were exposed to fluorescent light at 2000 lux for 10 or 60 min, then transferred to darkness for the indicated times. Mice were killed by cervical dislocation under anesthesia; retinoids in the retinas and RPE were separately extracted with hexane and analyzed by HPLC as described previously (Jin et al., 2009).
Immunoblot analysis
Proteins separated in a 10% or 12% polyacrylamide gel by SDS-PAGE were transferred to an Immobilon-P membrane (Millipore). The membrane was incubated in blocking buffer, primary antibody, and secondary antibody as described (Jin et al., 2005). Immunoblots were visualized with the enhanced ECL-Plus by scanning the membrane in an ImageQuant LAS4000 (GE Healthcare). The fluorescence intensity of each band was measured using ImageQuant TL software.
Electroretinography (ERG)
ERGs were recorded from the corneal surface of the eye, after pupil dilation with 1% tropicamide ophthalmic solution, using a silver-silver chloride wire electrode referenced to a subcutaneous electrode in the forehead. A needle electrode in the tail served as the ground. A drop of 2% methylcellulose was placed on the cornea to prevent corneal desiccation. Scotopic ERG responses were elicited with either short duration LED flashes or a Xenon strobe delivered in a Ganzfield dome (Espion, Diagnosys) with interstimulus intervals of 0.5–2 min, depending on the stimulus intensity. Flash intensities ranged from 0.0005 to 1000 cd-s/m2 in 16 steps. Two to four responses were averaged for each step. Responses were filtered using low-pass (0.15 Hz) and high-pass (100 Hz) filters. Intensity-response amplitude data were displayed on log-linear coordinates (SigmaPlot 11 software).
Light-induced retinal degeneration
129S2/Sv and Fatp4−/− mice were dark-adapted for three days. After dilation of the pupils under dim red light (Kodak Wratten 1A), mice were exposed to 15000 lux of white fluorescent light for 1.5 h and then kept in darkness for 5 days. Retinal degeneration was observed by spectral domain optical coherence tomography (SD-OCT) first and then analyzed by light microscopy.
SD-OCT
Retinas were imaged by SD-OCT along the vertical meridian (from superior to inferior) through the optic disc using a Heidelberg Spectralis HRA + OCT system (Heidelberg Engineering). Signal quality was greater than 20 db and scan speed was 40000 A-scans per second. Eye motion artifacts were eliminated by in-system eye tracking and at least 25 frames were averaged per B-scan to increase the signal to noise ratio (Zhou et al., 2011).
Light microscopy
Retinas were prepared as described previously (Knott et al., 2011). Care was taken to orient the eyecups so that sections were obtained through the vertical meridian to insure sampling of both the superior, green-sensitive cones and the inferior, blue-sensitive cones (Szél et al., 1992). One µm-thick plastic sections were contrasted with toluidine blue, cover-slipped, and imaged with a Nikon DS-Ri1 digital camera attached to a Nikon Optiphot-2 microscope using a Nikon Plan 40X oil-immersion lens and Nikon NIS Elements software v3.0.
Immunohistochemistry
Retinal cryosections, made as described previously (Jin et al., 2009), were incubated in a blocking solution containing DMEM, 10% FBS, 2% goat serum, and 0.1% Triton X-100 for 1 h, with primary antibodies at 4°C overnight, and with secondary antibodies at room temperature for 1 h, each followed by four washes with PBS containing 0.1% Tween-20. The primary antibodies used include affinity purified rabbit antibodies against FATP4 (Newberry et al., 2003), M-opsin, or S-opsin (Chemicon International) and mouse monoclonal antibodies against RPE65 (Millipore) or KDEL (Abcam). The secondary antibodies were Alexa Fluor 488 or 555 goat anti-rabbit or -mouse IgG (Invitrogen). Nuclei were labeled with 4',6-diamidino-2-phenylindole (DAPI). Fluorescent signals were captured with a Zeiss LSM-510 Meta laser confocal microscope with a 40X oil-immersion objective.
Quantitation of numbers and lengths of cone outer segments
We counted M-opsin positive outer segments (OS) along 500 μm linear regions in the superior retinas and S-opsin positive OS along 500 μm linear regions in the inferior retinas. In all cases, these 500 μm linear regions began at the optic nerve head and extended radially toward the superior or inferior peripheral retina. We also measured cone OS lengths in the same regions.
Quantitative reverse transcription-PCR (RT-PCR)
Total RNA was extracted from mouse RPE and retinas using the Absolutely RNA Miniprep kit (Stratagene), and was reverse-transcribed to cDNA using SuperScript III (Invitrogen). Quantitative PCR (qPCR) was performed on an iCylcer thermocycler (Bio-Rad ) using a two-step qRT-PCR kit with SYBR Green (Invitrogen) and primer sets specific for FATPs and 18S rRNA (Table 1). Three mice of each genotype were analyzed and all samples were run in duplicates. Starting templates were normalized after determining 18S rRNA Ct-values for each sample. Relative FATP mRNA levels were determined from the ΔCt values.
Table 1.
Primer pairs used in the quantitative RT-PCR
| primers | Sequence (5’-3’) | Primers | Sequence (5’-3’) |
|---|---|---|---|
| FATP1-F | gacaagctggatcaggcaag | FATP1-R | gtcttgcagacgatacgcaga |
| FATP2-F | gcctcggttcctgaggatac | FATP2-R | atgttctcagtcatgggcaca |
| FATP3-F | gagaacttgccaccgtatgc | FATP3-R | ggcccctatatcttggtcca |
| FATP4-F | gattctccctgttgctcctgt | FATP4-R | ccattgaagcaaacagcagg |
| FATP5-F | tgccacacctcatttcatcc | FATP5-R | acatctggcatcagactccg |
| FATP6-F | aaccaagtggtgacatctctgc | FATP6-R | tccataaagtaaagcgggtcag |
| 18S rRNA-F | tttgttggttttcggaactga | 18S rRNA-R | cgtttatggtcggaactacga |
DNA sequencing and database search
DNA sequencing was done using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI PRISM 3100 Genetic Analyzer. Database search was performed in the internet server http://www.ncbi.nlm.nih.gov/BLAST.
Statistical analysis
Statistical significance was determined with an unpaired, two-tailed Students t-test. P values less than 0.05 were considered to be statistically significant. Data are expressed as the mean ± standard deviation (SD) unless otherwise noted.
Results
Expression cloning of inhibitors for RPE65 isomerase
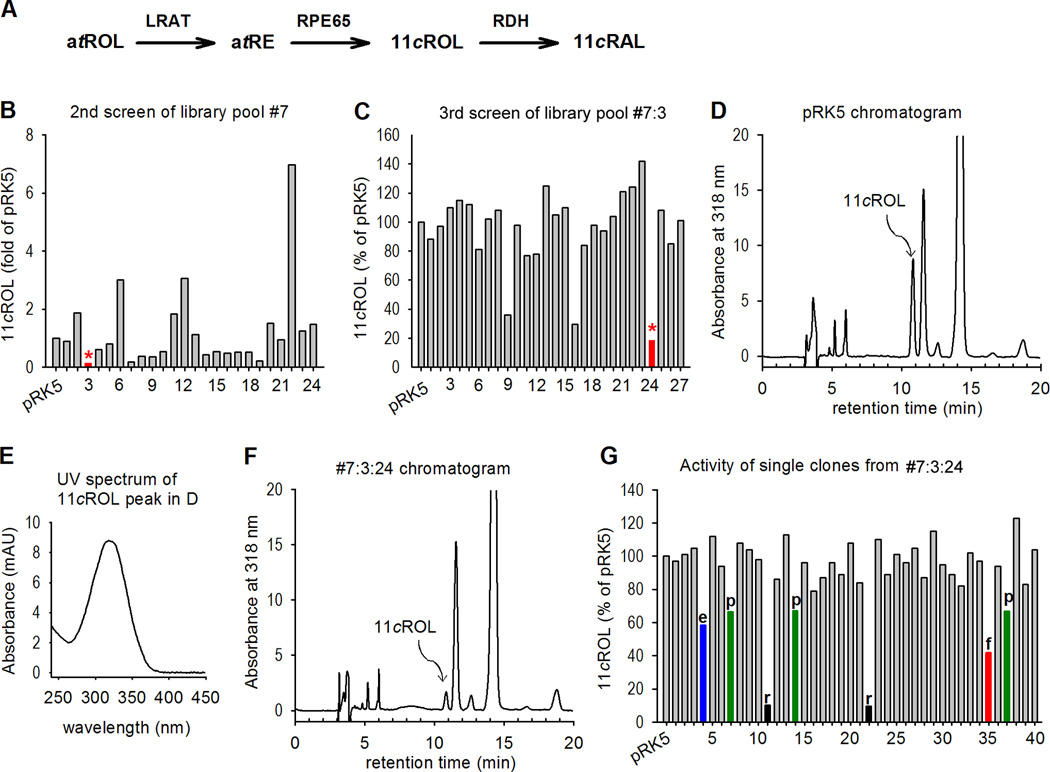
In our previous study (Jin et al., 2005), we observed that the amount of 11cROL in 293T-LRC cells transfected with bovine RPE expression library pool #7:3 was significantly smaller than that of control cells (Fig. 1B). One possible explanation for this small amount of 11cROL is that the pool #7:3 may contain a gene(s) that inhibits synthesis of 11cROL. To clone this unknown inhibitor(s), we split this pool #7:3, which contains approximately 650 clones, into 32 subpools each containing ~39 clones. We transfected 293T-LRC cells with plasmids from the 32 subpools plus pRPE65 for the next round of screening. We then measured the synthesis of 11cROL from atROL added to the cell media. Cells transfected with subpool #7:3:24 contained the smallest amount of 11cROL (Fig. 1C). We isolated 60 single clones from this subpool for the final screening. Cells transfected with 7 individual clones contained relatively smaller amounts of 11cROL (Fig. 1G). DNA sequencing and database search revealed that: clones #4 and #35 were identical to the bovine ELOVL1 cDNA (Gene ID: 540348) and the bovine FATP4/Slc27a4 cDNA (Gene ID: 781099), respectively; clones #11 and #22 were identical to the bovine RDH5 (Gene ID: 281448); and clones #7, #14, and #37 were identical to the bovine 26S proteasome non-ATPase regulatory subunit-13 (PSMD13, Gene ID: 513525).
Figure 1. Expression screening for inhibitor of RPE65.
(A) The visual cycle reactions taking place in RPE. LRAT esterifies all-trans retinol (atROL) from photoreceptors and blood to all-trans retinyl esters (atRE), such as all-trans retinyl palmitate (atRP); RPE65 isomerohydrolyzes atRP to 11-cis retinol (11cROL), which is then oxidized by retinol dehydrogenase (RDH) to 11-cis retinaldehyde (11cRAL). (B) Secondary screening of 30 subpools (24 shown) from a bovine RPE expression library pool #7 (Jin et al., 2005). Amounts of 11cROL synthesized from atROL added into the media of 293T-LRC cells transfected with the indicated library pools were analyzed by HPLC. Cells transfected with subpool #3 (*) contained the least amount of 11cROL. (C) Tertiary screening of 32 subpools (27 shown) of pool #7:3. Isomerase assays were performed as in (B). Subpool #7:3:24 (*) was selected for the final screening. (D) HPLC chromatogram of retinoids extracted from the control cells transfected with pRPE65 and pRK5 (mock) plasmids. The peak corresponding to 11cROL is indicated. (E) UV spectrum acquired from the 11cROL peak in (D). (F) HPLC chromatogram of retinoids extracted from the cells transfected with subpool #7:3:24. (G) Isomerase assays of 60 single clones (40 shown) from subpool #7:3:24. Clones for ELOVL1 (e), RDH5 (r), FATP4 (f), and PSMD13 (p) are indicated.
FATP4 and ELOVL1 had no effect on oxidation of 11cROL and esterification of atROL
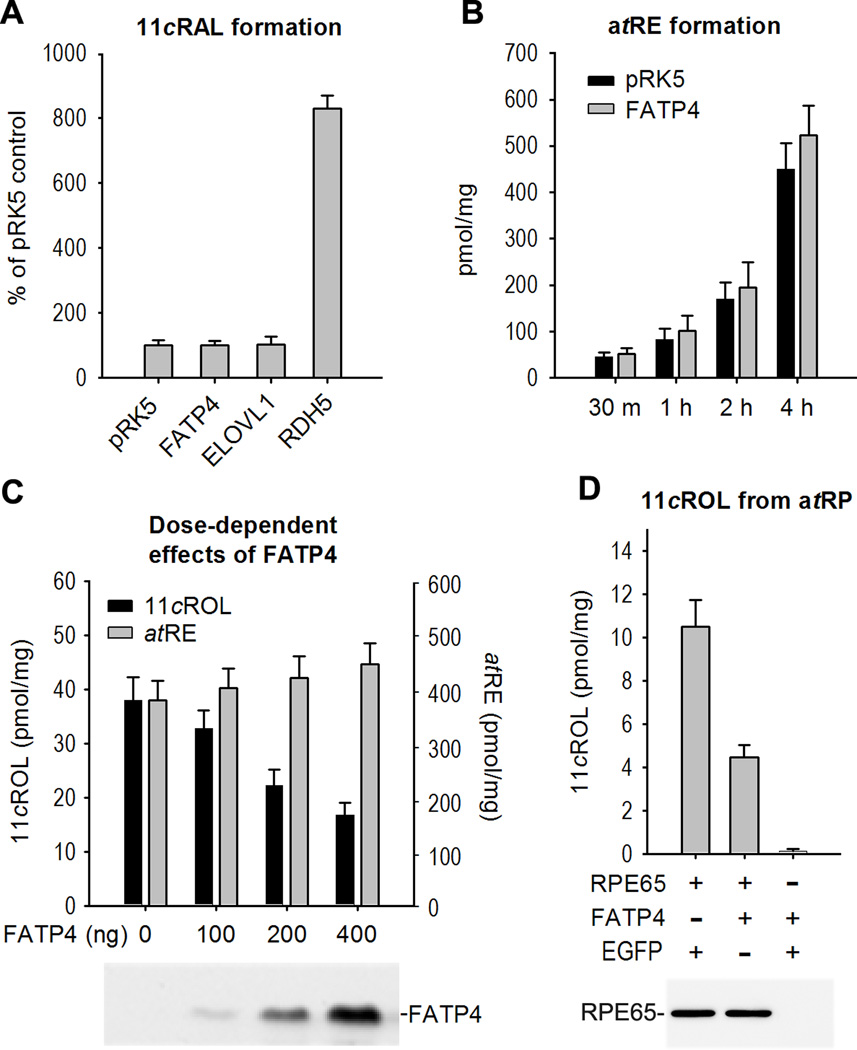
Since RDH5 has been shown to catalyze oxidation of 11cROL to 11cRAL (Simon et al., 1995), we measured the amounts of 11cRAL and 11cROL in 293T-LRC cells cotransfected with pRPE65 plus either RDH5, ELOVL1, FATP4, or pRK5 (control) plasmids. Consistent with the screening result (Fig. 1G), the amount of 11cROL in the RDH5-cotransfected cells was approximately 10% of those in the pRK5-cotransfected control cells. In contrast, the amount of 11cRAL in the RDH5-cotransfected cells was nearly 9-fold higher than that in the control cells (Fig. 2A), indicating that the small amount of 11cROL in the RDH5-cotransfected cells was due to oxidation of 11cROL to 11cRAL by RDH5. On the other hand, the amounts of 11cRAL in the ELOVL1- or FATP4-cotransfected cells were similar to those in the pRK5-cotransfected control cells (Fig. 2A), indicating that neither ELOVL1 nor FATP4 catalyzed oxidation of 11cROL to 11cRAL.
Figure 2. FATP4 had no effect on synthesis of atRE and 11cRAL but inhibited synthesis of 11cROL.
(A) RDH5, but not FATP4 or ELOVL1, oxidized 11cROL to 11cRAL. 293T-LRC cells transfected with the indicated plasmids were incubated with atROL. 11cRAL in the cells was converted to syn– and anti-retinaloxime isomers by hydroxylamine treatment, and was measured by HPLC. (B) LRAT assay showing synthesis of atRE from atROL incubated for the indicated times with the cells transfected with pRK5 (control) or FATP4 plasmid. (C) Dose dependent inhibition of 11cROL synthesis by FATP4 in living cells. Synthesis of 11cROL (left Y-axis) and atRE (right Y-axis) from atROL in 293T-LRC cells transfected with the indicated amounts of FATP4 plasmid were measured by HPLC. A representative immunoblot shows expression levels of FATP4 in the transfected cells. (D) In vitro isomerase assay showing synthesis of 11cROL from atRP substrate incubated with homogenates of Sf9 cells infected with the indicated recombinant baculoviruses. A representative immunoblot shows similar expression levels of RPE65 in the baculovirus-infected cells. All error bars are standard deviation (SD, n = 4).
Although atRP is a proven substrate of RPE65, its strong hydrophobicity prompted us to add atROL into the media of 293T-LRC cells as a substrate. Therefore, synthesis of 11cROL in this assay consisted of two enzymatic reactions: the first was synthesis of atREs, such as atRP, from atROL by LRAT; and the second was the isomerization of atRP to 11cROL by RPE65 (Fig. 1A). We therefore tested if FATP4 inhibited LRAT activity. As shown in Fig. 2B, the amounts of atRE in the FATP4-transfected cells were similar to those in the control cells transfected with pRK5 mock plasmid. We then analyzed the dose-dependent effect of FATP4 on the synthesis of 11cROL. As shown in Fig. 2C, synthesis of 11cROL in the cells decreased as FATP4 expression levels increased (Fig. 2C, left Y-axis), while synthesis of atRE was not significantly changed as FATP4 increased (Fig. 2C, right Y-axis, P > 0.05). These results suggest that FATP4 inhibited 11cROL synthesis catalyzed by RPE65. To confirm this result, we performed an in vitro isomerase assay using atRP as the substrate. Homogenates of Sf9 cells expressing RPE65 plus EGFP synthesized approximately 11-pmol of 11cROL from atRP, whereas homogenates of Sf9 cells expressing RPE65 plus FATP4 synthesized only ~4.3-pmol of 11cROL (Fig. 2D). Expression levels of RPE65 in these two groups of cells were similar (Fig. 2D), indicating that FATP4 inhibited RPE65-catalyzed synthesis of 11cROL.
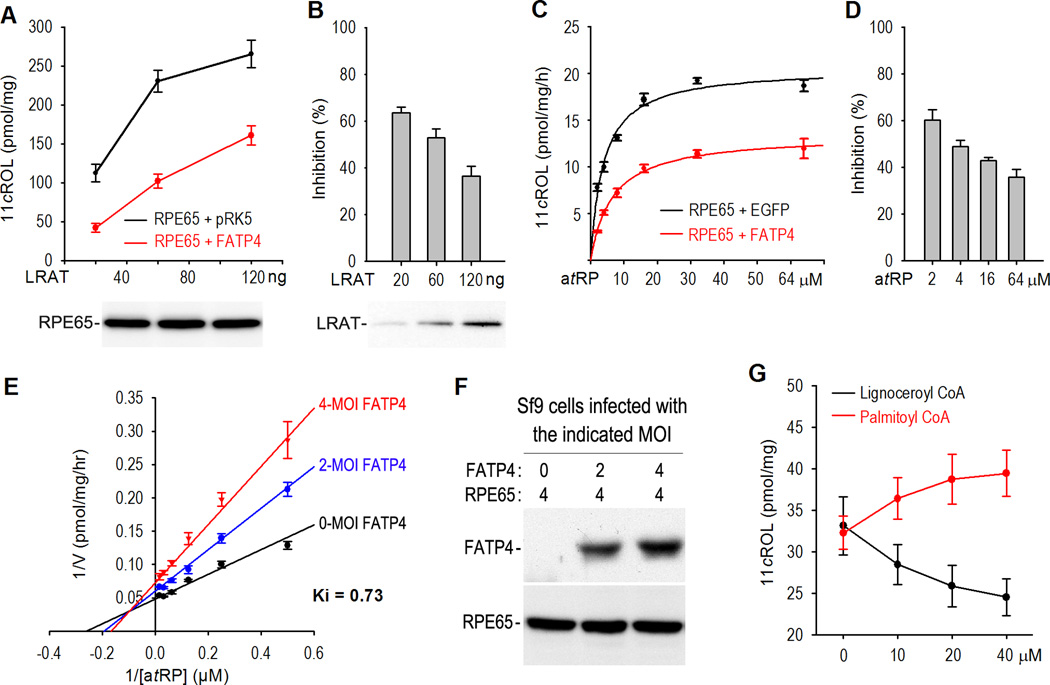
Substrate competition and VLCFA-ACS product are involved in the FATP4-mediated inhibition of RPE65
To define the mechanisms by which FATP4 inhibits synthesis of 11cROL, we assayed isomerase activity in 293T-C cells transfected with a combination of a series of LRAT amounts plus a constant amount of RPE65 and either FATP4 or pRK5. Synthesis of 11cROL in the cells expressing RPE65 and FATP4 was significantly lower compared to that of the control cells (Fig. 3A). Synthesis of 11cROL was reduced ~60% by FATP4 in the cells transfected with 20 ng of LRAT, whereas it was reduced only ~35% by FATP4 in the cells transfected with 120 ng of LRAT (Fig. 3B), suggesting that LRAT suppressed FATP4-mediated inhibition of RPE65. This result can be explained by a competition between FATP4 and RPE65 for RPE65’s substrate, atRP, which is synthesized by LRAT. To test this possibility, we performed an in vitro assay using a series of increasing amounts of atRP and Sf9 cells expressing RPE65 plus EGFP or FATP4. As expected, the inhibition of 11cROL synthesis by FATP4 was significantly reduced as atRP increased (Fig. 3C, D). These results suggest that FATP4 competes with RPE65 for RPE65’s substrate.
Figure 3. FATP4 functions as a mixed-type inhibitor of RPE65.
(A) Synthesis of 11cROL from atROL added into the media of 293T-C cells transfected with the indicated amounts of LRAT and constant amounts of RPE65 plus FATP4 or pRK5. A representative immunoblot shows similar expression levels of RPE65 in the transfected cells. (B) Relative inhibition rate of 11cROL synthesis by FATP4 in (A). Note that the inhibition rate was reduced as the amount of transfected LRAT increased. The immunoblot shows different expression levels of LRAT in the transfected cells. (C) Synthesis of 11cROL from varying concentrations of the atRP substrate incubated with homogenates of Sf9 cells expressing RPE65 plus EGFP or FATP4. (D) Relative inhibition rate of RPE65 activity by FATP4 in (C). Note that the inhibition rate was reduced as the concentration of atRP substrate increased. (E) Lineweaver-Burk plot for a dataset from a series of isomerase assays using varying concentrations of the atRP substrate and Sf9 cells infected with RPE65 baculovirus (MOI=4) and the indicated deferent MOI of FATP4 baculovirus. (F) Immunoblots show expression levels of FATP4 and RPE65 in the baculovirus-infected Sf9 cells that were used for isomerase assays in (E). (G) Effects of long-chain (palmitoyl; 16:0) and very long-chain (lignoceroyl; 24:0) fatty acyl-CoA on the synthesis of 11cROL in an in vitro isomerase assay. Error bars show SD (n = 3).
To further analyze the inhibitory mechanisms, we measured the inhibition rate using Sf9 cell homogenates containing different amounts of FATP4 (Fig. 3F) in the presence of varying amounts of atRP substrate. Unexpectedly, the Lineweaver-Burk plot for the assay data did not fit a typical competitive inhibition model (Fig. 3E). Instead, the data showed that FATP4 functions as a mixed type inhibitor of RPE65.
Both FATP4 and ELOVL1 have very long-chain fatty acid acyl-CoA synthetase (VLCFA-ACS) activity, with the highest activity toward saturated VLCFAs (22:0–24:0). We therefore tested if their products can inhibit isomerase activity. As shown in Fig. 3G, palmitoyl (C16:0)-CoA promoted synthesis of 11cROL, whereas lignoceroyl (C24:0)-CoA inhibited synthesis of 11cROL, suggesting that products of FATP4 and ELOVL1 can negatively regulate synthesis of 11cROL catalyzed by RPE65.
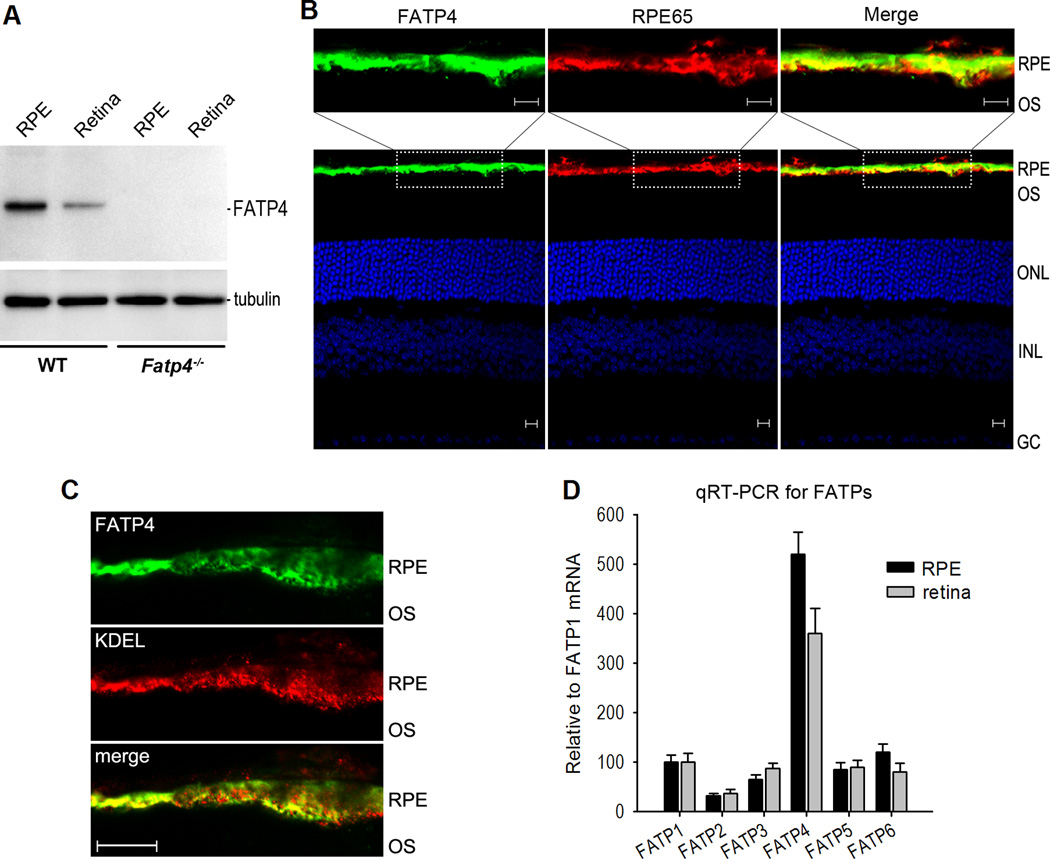
FATP4 is the predominant FATP in RPE
FATP4 is one of six-members (FATP1–6) of an integral transmembrane protein family, each of which shows different tissue expression patterns. To begin to define the roles of FATP4 in regulation of the visual cycle in vivo, we analyzed expression of FATP4 in the retina and RPE. Immunoblot analysis showed that FATP4 was expressed strongly in the wild-type RPE but very weakly in the retina (Fig. 4A). In contrast, FATP4 was undetectable in the Fatp4−/− RPE and retina (Fig. 4A). Immunohistochemistry confirmed that FATP4 was predominantly expressed in the wild-type mouse RPE (Fig. 4B). Moreover, quantitative reverse transcription PCR (qRT-PCR) demonstrated that FATP4 is the predominant FATP in RPE (Fig. 4D).
Figure 4. FATP4 is the predominant FATP in RPE.
(A) Immunoblot analysis of FATP4 and tubulin in retinas and RPE from 129S2/Sv (WT) and Fatp4−/− mice. (B) Immunohistochemistry showing FATP4 in the WT mouse RPE and its colocalization with RPE65. RPE, outer segments (OS) of photoreceptors, outer nuclear layer (ONL), inner nuclear layer (INL) and ganglion cell (GC) layer are indicated. Scale bars denote 10 µm. (C) Immunohistochemistry showing colocalization of FATP4 with KDEL, an ER marker antibody, in the mouse RPE. Scale bar denotes 10 µm. (D) Quantitative RT-PCR showing relative expression levels of six FATP family members in the mouse RPE and retina. Relative FATP mRNA levels are shown as the percent of FATP1 mRNA level. Error bars represent SD (n = 3).
To inhibit RPE65 in a competitive fashion, at least some FATP4 and RPE65 proteins should colocalize in RPE. We therefore tested this possibility by immunohistochemistry using antibodies against FATP4 and RPE65, and found that a majority of the FATP4 was co-localized with RPE65 in the wild-type RPE (Fig. 4B). A previous study showed that RPE65 is associated with the smooth endoplasmic reticulum (sER) (Sagara and Hirosawa, 1991). FATP4 has an ER localization signal and localizes to the ER in cultured cells (Milger et al., 2006). We therefore tested whether FATP4 localizes to the ER of mouse RPE. Immunocytochemistry and confocal microscopy revealed that a majority of FATP4 colocalized with an ER marker (Fig. 4C), indicating that FATP4 is associated with the ER.
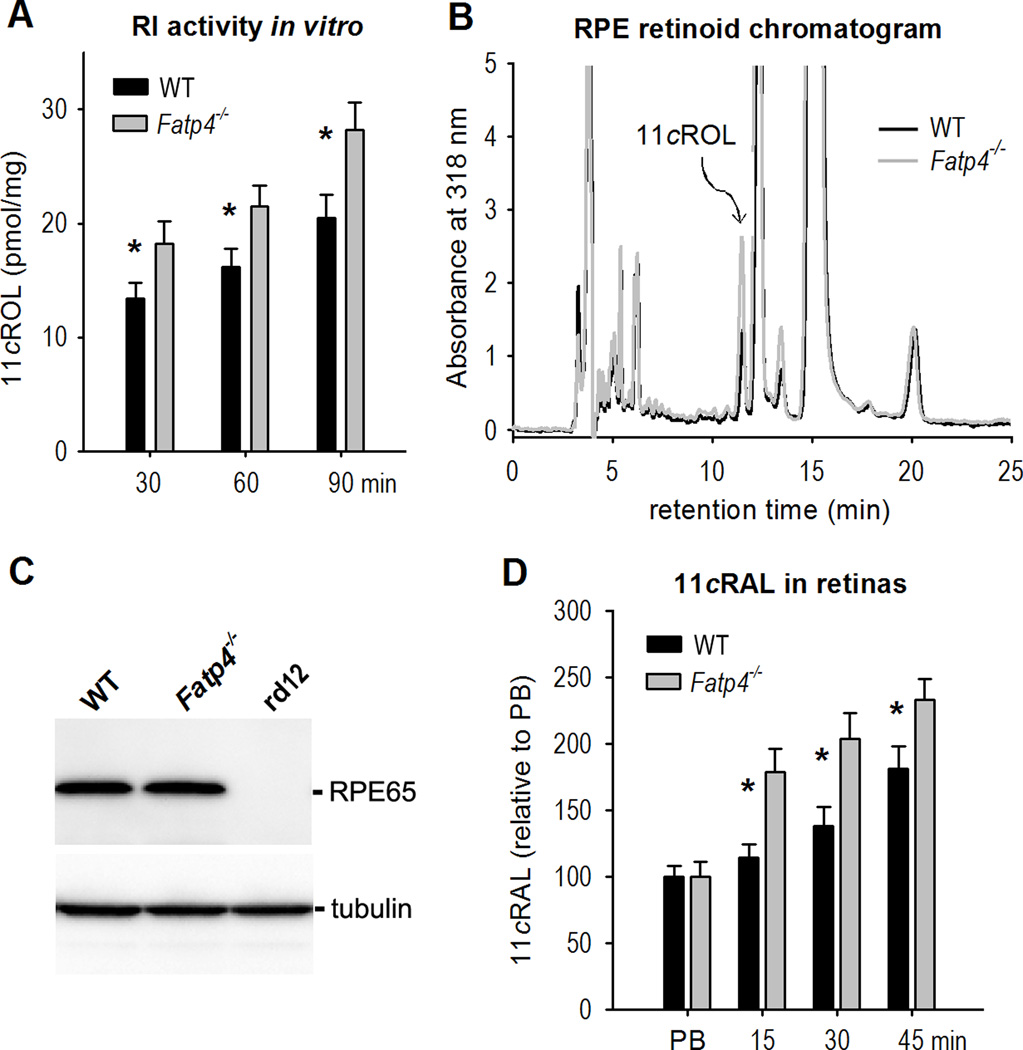
Hyper-isomerase activity and fast 11cRAL regeneration in Fatp4−/− Mice
To test if FATP4 inhibits RPE65 function in vivo, we compared synthesis of 11cROL in RPE homogenates from wild-type and Fatp4−/− mice. As shown in Fig. 5A and 5B, the amount of 11cROL synthesized by Fatp4−/− RPE was 24–30% higher than that synthesized by wild-type RPE. We tested if this higher isomerase activity in Fatp4−/− RPE was due to up-regulation of RPE65. Immunoblot analysis showed that the expression level of RPE65 in Fatp4−/− RPE was similar to that in the wild-type RPE (Fig. 5C), indicating that FATP4 inhibited synthesis of 11cROL without impacting RPE65 expression. To determine if FATP4 delays the regeneration rate of 11cRAL, we measured the contents of 11cRAL in the retinas of wild-type and Fatp4−/− mice kept in darkness for different times after photo-bleaching their visual pigments. The 11cRAL level in Fatp4−/− retinas increased more quickly compared to that in wild-type (Fig. 5D), indicating that FATP4 slows down the visual cycle.
Figure 5. Hyper-isomerase activity and faster regeneration of 11cRAL in Fatp4−/− mice.
(A) Retinoid isomerase (RI) activities in WT and Fatp4−/− RPE cells. Synthesis of 11cROL by WT and Fatp4−/− RPE homogenates during the indicated incubation times was measured by HPLC. Asterisks indicate statistically significant differences (P < 0.05). Error bars show SD (n = 4). (B) Representative HPLC chromatograms of retinoids from WT and Fatp4−/− RPE homogenates assayed for 90 minutes in (A). The peaks corresponding to 11cROL are marked (arrow). (C) Immunoblot analysis showing similar expression levels of RPE65 in WT and Fatp4−/− RPE cells. RPE cells from RPE65-deficient rd12 mice were used as a negative control. (D) Relative contents of 11cRAL in the retinas from mice immediately after photo-bleaching (PB) of the visual pigment, or from mice kept in darkness for the indicated times after PB.
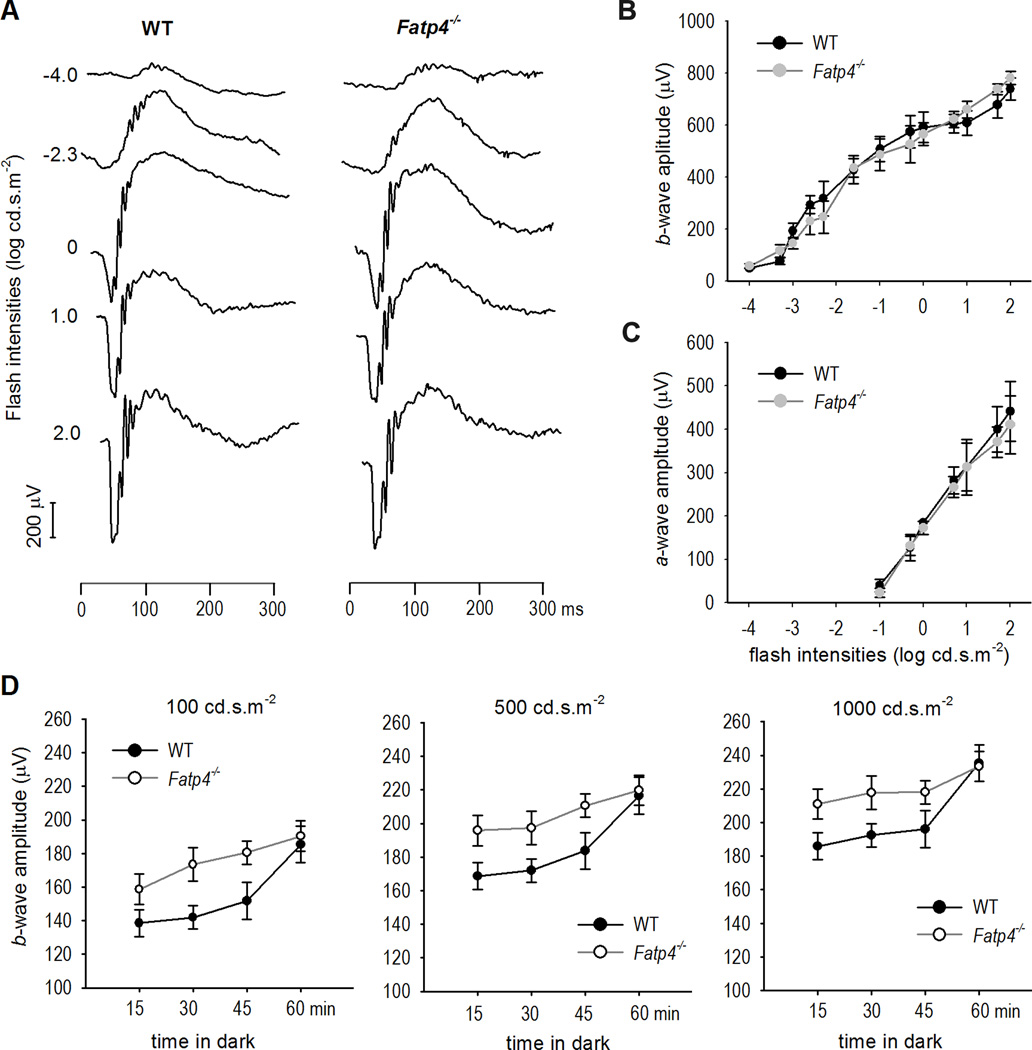
Fast dark adaptation in Fatp4−/− Mice
To confirm the result shown in Fig. 5, we recorded the recovery rate of rod light sensitivity in Fatp4−/− mice. Overnight dark-adapted C57BL/6J and Fatp4−/− mice displayed similar ERG responses to a series of flash stimuli (Fig. 6A–C). However, Fatp4−/− mice kept in darkness for short times after photobleaching displayed higher b-wave amplitudes compared to wild-type mice (Fig. 6D). This result is consistent with the finding that the b-wave recovery in BALB/c mice with a higher expression level of RPE65 was faster than that in c2J mice with a lower expression level of RPE65 (Nusinowitz et al., 2003), indicating that FATP4 negatively regulates the recovery rate of rod function.
Figure 6. Faster recovery of rod light sensitivity in Fatp4−/− mice.
(A) Representative raw scotopic ERG tracings show similar responses of overnight dark-adapted C57BL/6J (WT) and Fatp4−/− mice to the indicated flash intensities. (B & C) Similar a- and b-wave amplitudes observed in overnight dark-adapted WT and Fatp4−/− mice. Error bars show SD (n = 5). (D) ERG responses to the indicated flash intensities in WT and Fatp4−/− mice kept in darkness for the indicated times after photo-bleaching (PB). Error bars show SD (n = 5).
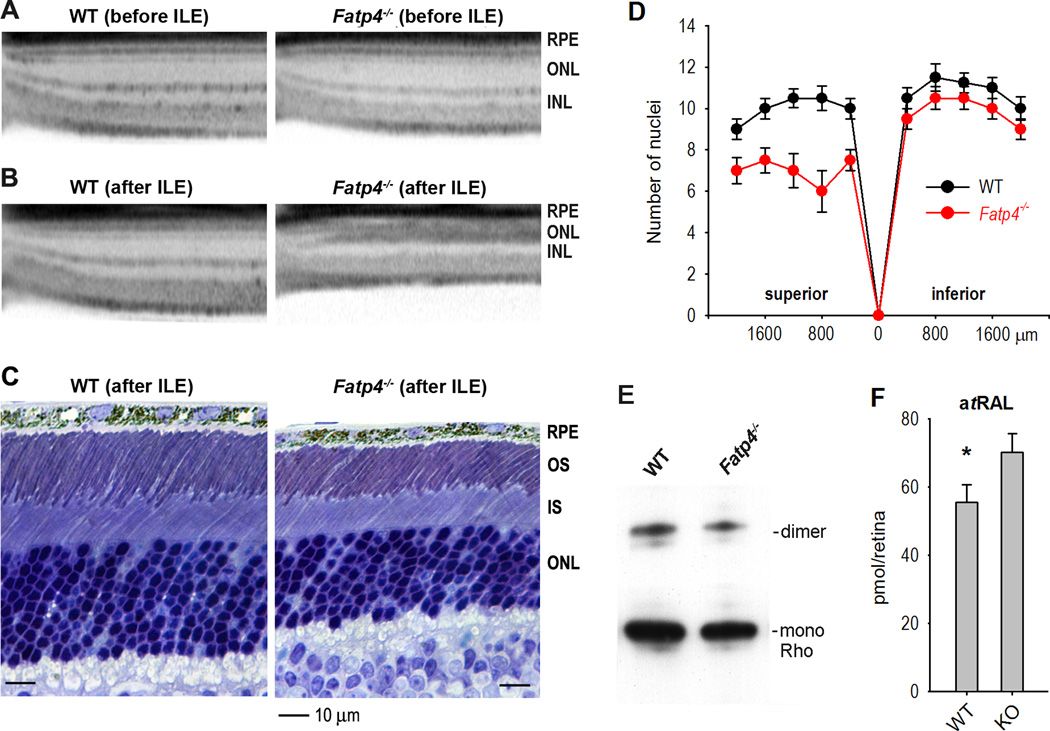
FATP4 is necessary to prevent rod and cone degeneration induced by intense light
Previous studies have shown that RPE65 regulates light-induced retina degeneration (Danciger et al., 2000; Wenzel et al., 2001). We therefore tested if Fatp4−/− mice exhibit higher susceptibility to intense light. Before exposing to intense light, 129S2/Sv and Fatp4−/− mice showed similar retinal structure and thickness in SD-OCT imaging (Fig. 7A). After exposing to 15000 lux light for 1.5 h, however, Fatp4−/− mice, but not WT mice, showed severe retinal (mainly rod) degeneration by both SD-OCT and light microscopy (Fig. 7B, C). Superior outer nuclear layer in Fatp4−/− retinas were reduced in thickness to 6–8 nuclei versus 9–11 nuclei in wild-type retinas (Fig. 7D). Immunoblot analysis confirmed that rhodopsin content was reduced in Fatp4−/− mice after intense light exposure (Fig. 7E). Since atRAL is a cytotoxic effector in light-induced retinal degeneration (Maeda et al., 2009), we measured the content of atRAL in the retinas. As shown in Fig. 7F, retinas of Fatp4−/− mice exposed to 2000 lux light for 1 h contained ~26% higher atRAL compared to wild-type mice under the same lighting condition.
Figure 7. FATP4 prevents degeneration of rod photoreceptors induced by intense light.
(A) Spectral domain optical coherence tomography (SD-OCT) shows similar structures of superior retinas in 129S2/Sv (WT) and Fatp4−/− mice before intense light exposure (ILE). RPE, outer nuclear layer (ONL), and inner nuclear layer (INL) are indicated. (B & C) OCT and light microscopy show retinal (mainly rods) degeneration in the superior retina of Fatp4−/− mice exposed to 15000 lux for 1.5 h. (D) Retinal outer nuclear layer thickness in WT and Fatp4−/− mice exposed to intense light. Numbers on the X-axis indicate distance from optic nerve head. Error bars show SD (n = 3). (E) Immunoblots for rhodopsin (Rho) in WT and Fatp4−/− retinas exposed to intense light. Bands corresponding to Rho dimers and monomers are indicated. (F) Contents of all-trans retinaldehyde (atRAL) in the retinas of WT and Fatp4−/− mice exposed to 2000 lux light for 1 h. Asterisk indicates a statistically significant difference (P < 0.05). Error bars show SD (n = 3).
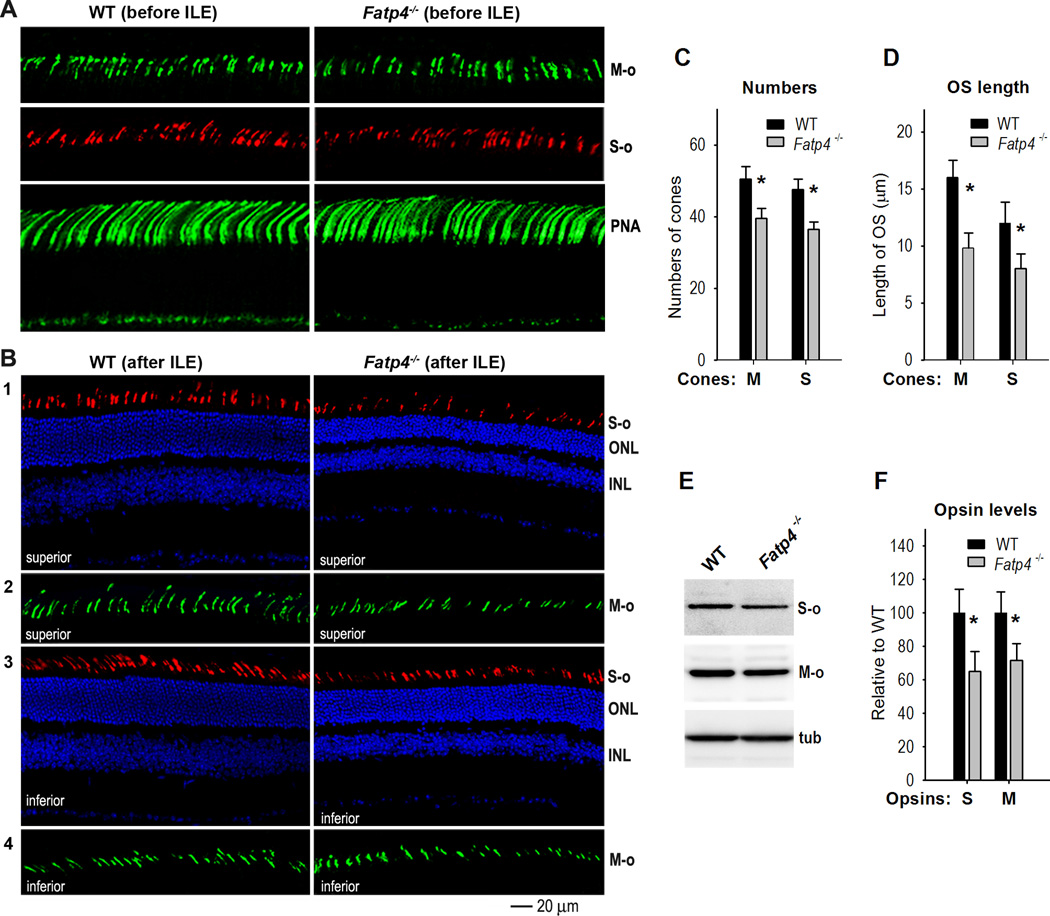
Previous studies have shown that intense light triggers cone photoreceptor death in rat (Cortina et al., 2003). To test if cones also degenerated in Fatp4−/− mice exposed to intense light, we stained retinal sections with antibodies against M-opsin or S-opsin. Before intense light exposure, 129S2/Sv and Fatp4−/− mice had similar numbers and structure of M- and S-cone photoreceptor outer segments (Fig. 8A). Peanut agglutinin (PNA)-staining confirmed that numbers and length of cone outer segment sheathes in Fatp4−/− retinas were similar to those in wild-type retina (Fig. 8A). After exposing to intense light, however, Fatp4−/− mice had a significantly smaller number of M- and S-opsin positive outer segments compared to wild-type mice (Fig. 8B, C). Lengths of M- and S-cone outer segments in Fatp4−/− mice were also shorter than those of wild-type mice (Fig. 8B, D). Consistent with these results, M- and S-opsin levels in Fatp4−/− retinas were ~70% of those in wild-type retinas (Fig. 8E, F).
Figure 8. FATP4 prevents degeneration of cone photoreceptors induced by intense light.
(A) Immunohistochemistry with an antibody against M-opsin (M-o) or S-opsin (S-o) shows similar lengths and numbers of cone outer segments in 129S2/Sv (WT) and Fatp4−/− superior retinas before intense light exposure (ILE). Fluorescein-peanut agglutinin (PNA) staining confirms similar lengths and numbers of cone photoreceptor matrix sheathes in WT and Fatp4−/− superior retinas. (B) Degeneration of M- and S-cone photoreceptors in Fatp4−/− mice after intense light exposure (ILE). Panels 1 and 2: immunostaining of S- and M-opsin in the superior retinas of WT and Fatp4−/− mice. Panels 3 and 4: immunostaining of S- and M-opsin in the inferior retinas of WT and Fatp4−/− mice. ONL, outer nuclear layer; INL, inner nuclear layer. (C) Average numbers of M- or S-opsin positive cells in a superior retinal region from the optic nerve to 500 µm. Asterisks indicate significant differences (P < 0.05). Error bars show SD (n = 4). (D) Average length of M- and S-cone outer segments in the same retina regions described in (C). (E) Immunoblot analysis of M- and S-opsin in WT and Fatp4−/− retinas exposed to intense light. Tubulin (tub) was utilized to normalize sample loading. (F) Densitometry analysis of the immunoblots in (E) to quantitate relative contents of M- and S-opsin in WT and Fatp4−/− retinas. Asterisks indicate significant differences (P < 0.05). Error bars show SD (n = 3).
Discussion
The purpose of this study was to identify and characterize previously unknown inhibitor(s) of RPE65, which controls the rate-limiting step of the visual cycle and retinal susceptibility to light-induced degeneration. Through an unbiased screening of a bovine RPE cDNA library we isolated FATP4, ELOVL1, PSMD13, and RDH5 as candidate negative regulators of RPE65 (Fig. 1G).
RDH5 is a stereospecific RDH that catalyzes oxidation of 11cROL to 11cRAL (Simon et al., 1995). As predicted, the reduced levels of 11cROL in the RDH5-transfected cells (Fig. 1G) was due to oxidation of 11cROL to 11cRAL by RDH5 (Fig. 2A). We therefore conclude that RDH5 is not an inhibitor of RPE65. Although RDH5 is not an inhibitor of RPE65, our results help to explain why the isomerase activity in bovine RPE microsomes is very low. RDH5 is a membrane-bound protein. A majority of bovine RDH5 is associated with smooth ER in RPE (Simon et al., 1999), indicating that RDH5 should be present in RPE microsomes. This subcellular localization suggests that the low amount of 11cROL in the isomerase assay of bovine RPE microsomes (Winston and Rando, 1998) is partially due to oxidation of 11cROL to 11cRAL by RDH5.
PSMD13 is a non-ATPase regulatory subunit of proteasome whose main function is to degrade proteins. PSMD13 promoted degradation of RPE65 (unpublished observation). Therefore, PSMD13 might reduce the synthesis of 11cROL by promoting degradation of RPE65.
Both FATP4 and ELOVL1 are involved in the metabolism of saturated and monounsaturated LCFA and VLCFA. FATP4 is a 72-kDa protein with an N-terminal transmembrane domain, an ER localization signal, and ATP/AMP and FATP motifs. It mediates uptake of LCFA and VLCFA, and plays a central role in epidermal barrier formation (Moulson et al., 2003). FATP4 exhibits low palmitoyl-CoA and high lignoceroyl-CoA synthetase activities (Hall et al., 2005). ELOVL1 is a 32-kDa protein with 5 putative membrane-spanning domains and a C-terminal ER retrieval signal (Tvrdik et al., 2000). Like other elongases, ELOVL1 catalysis the first of four steps in the VLCFA elongation cycle. It exhibits activity toward saturated C18 to C26 acyl-CoA substrates, with the highest activity towards C22:0 acyl-CoA. It is also essential for synthesis of C24:0 and C24:1 sphingolipids.
Similar to the control cells transfected with pRK5, cells transfected with FATP4 or ELOVL1 plasmids synthesized trace amounts of 11cRAL (Fig. 2A), suggesting that these two proteins did not catalyze oxidation of 11cROL to 11cRAL. Also, these two proteins did not inhibit synthesis of atRE (Fig. 2B, C). However, synthesis of 11cROL was significantly inhibited by these two proteins (Fig. 1G), suggesting that the action points of FATP4 and ELOVL1 are on the RPE65-catalyzed all-trans to 11-cis isomerization. The in vitro isomerase assay with atRP substrate confirmed that FATP4 inhibited isomerization of atRP to 11cROL (Fig. 3), indicating that FATP4 is a negative regulator of RPE65.
Reportedly, the dissociation constants (KD) of RPE65 for atRP, farnesyl palmitate, and all-trans retinyl acetate (atRAc) are 47 ± 3 nM, 63 ± 1 nM, and 1300 ± 42 nM, respectively (Maiti et al., 2005), indicating that the palmitic acyl (PA) moiety of atRP and farnesyl palmitate are important for their binding with RPE65. FATP4 can bind PA and palmitoyl-CoA (Stahl et al., 1999), suggesting that FATP4 may compete with RPE65 for binding to atRP, the substrate of RPE65. This explanation is consistent with the results presented in Fig. 3A–D: the FATP4-mediated inhibition of 11cROL synthesis was reduced as the amounts of LRAT and atRP increased. On the other hand, the Lineweaver-Burk plot for the dataset obtained from the kinetic assay (Fig. 3E) indicated that FATP4 was a mixed type inhibitor of RPE65.
Although both FATP4 and ELOVL1 could inhibit synthesis of 11cROL (Fig. 1G), these two proteins share only 11% amino acid homology. The common biochemical feature of FATP4 and ELOVL1 is their VLCFA-CoA synthetase activities, suggesting that VLCFA-CoA may inhibit RPE65 activity. In support of this hypothesis, lignoceroyl-CoA inhibited synthesis of 11cROL (Fig. 3G). Lignoceroyl-CoA itself or its all-trans retinyl ester (all-trans retinyl lignocerate) synthesized by acyl CoA:retinol acyltransferase (Kaschula et al., 2006) may compete with atRP for binding to RPE65. Since the fatty acyl moiety of all-trans retinyl lignocerate is much longer than that of atRP, all-trans retinyl lignocerate is not a preferred substrate for RPE65, but it could compete with atRP for the hydrophobic pocket containing the catalytic iron atom in RPE65 (Kiser et al., 2009). Therefore, FATP4-mediated inhibition of RPE65 may include two mechanisms: 1) FATP4 competes with RPE65 for the substrate of RPE65; and 2) all-trans retinyl lignocerate and/or lignoceroyl-CoA synthesized by FATP4 competes with atRP for the hydrophobic pocket of RPE65. In any event, our findings suggest that FATP4, and ELOVL1, and VLCFA comprise regulatory elements of the visual cycle and could be new therapeutic targets for degenerative diseases associated with an aberrant visual cycle.
Still, the significantly lower activity of the retinoid isomerase in RPE microsomes cannot be explained completely by the results presented above, and may involve the following mechanisms. First, all-trans to 11-cis isomerization is a thermodynamically uphill reaction (Deigner et al., 1989). Second, other inhibitory factors are present in RPE microsomes. The FATP and ELOVL families contain 6 and 7 members, respectively. A previous study showed that FATP1 inhibited synthesis of 11cROL (Guignard et al., 2010). Although the effects of other family members on the synthesis of 11cROL have not yet been studied, their similarities in subcellular location and enzymatic nature suggest that at least some of the members may inhibit synthesis of 11cROL. Lastly, a positive regulator(s) may be lost from RPE microsomes.Lopes et al. (2011) recently found that MYO7A is required for normal localization and function of RPE65. MYO7A is a cytoplasmic protein and thus may be not present in the microsome fraction, although it is required for RPE65 function.
A spontaneous mutant mouse line called wrinkle free (wrfr) was identified by positional cloning to have a retrotransposon insertion in the Fatp4 gene (Moulson et al., 2003). The wrfr mice show neonatal death due to severe skin defects and asphyxia.Moulson et al. (2007) rescued the lethality by expressing FATP4 solely in keratinocytes via a transgene. FATP4 was not detectable in the rescued transgenic mouse retina and RPE (Fig. 4A), indicating that this transgenic mouse line (shown as Fatp4−/− in this study) is a good model for studying the role of FATP4 in vision. The visual cycle phenotypes, including the hyper-isomerase activity, the faster chromophore regeneration and dark-adaptation rates in Fatp4−/− mice (Fig. 5, 6) demonstrate that FATP4 negatively regulates the visual cycle by inhibiting RPE65. These results also indicate that our library screening is a useful method to identify new regulator(s) of the visual cycle.
The rod and cone photoreceptors in Fatp4−/− mice exhibited significantly increased susceptibility to light-induced degeneration (Fig. 7, 8), indicating that FATP4 is essential for preventing retinal degeneration induced by light damage. Distinct susceptibilities of retinas to light damage were found in mice with a mutation in the Rpe65 gene (Danciger et al., 2000; Grimm et al., 2000; Wenzel et al., 2001) and has identified rhodopsin, whose regeneration rate depends on the visual cycle rate, as the mediator of light-induced photoreceptor apoptosis (Grimm et al., 2000). The 129/Ola strain with the Leu450 allele of Rpe65 exhibits higher susceptibility to light damage compared to C57BL/6J and B6;129S (N2) mice harboring the Met450 allele of Rpe65 (Wenzel et al., 2001). This hyper-susceptibility in the 129/Ola strain reflects its higher expression of RPE65 (Wenzel et al., 2001). Conversely, many factors or treatments that reduce RPE65 activity or expression reduced photoreceptor degeneration induced by intense light (Sieving et al., 2001; Maeda et al., 2006; Lopes et al., 2011). Based on these previous studies and our results, we conclude that the increased susceptibility of Fatp4−/− photoreceptors to light-induced degeneration was at least partially due to the increased activity of RPE65, which caused higher accumulation of cytotoxic atRAL in the retina (Fig. 7F). Approximately 30% longer photoreceptor outer segments in the superior versus inferior retinas (Battelle and LaVail, 1978) may be associated with the increased susceptibility of the superior retina to light damage (Fig. 8B), because longer outer segments would contain more rhodopsin that has been shown to mediate retinal degeneration caused by intense light.
Recently, mutations in the human FATP4 gene have been identified in patients with ichthyosis prematurity syndrome (Klar et al., 2009; Sobol et al., 2011), a recessive disorder characterized by premature birth, thick caseous desquamating epidermis, and neonatal asphyxia (Bygum et al., 2008). These reports did not mention visual problems in the patients. The lack of significant vision impairment could be due to 1) the functional redundancy of FATP family members, 2) the relatively strong resistance of human retina to light damage, and 3) the patients described are newborn or children. Since A2E accumulates in RPE with age, and since the Fatp4−/− retina contained a higher content of atRAL (Fig. 7F), it is important to investigate whether FATP4 mutations induce age-related vision impairment and retinal degeneration.
Acknowledgements
This work was supported by NIH grants EY021208 (M.J.) and AR049269 (J.H.M.), and by an intramural fund from the LSU Health Sciences Center (M.J.). We thank Drs. Nicholas O. Davidson and Elizabeth P. Newberry for the FATP4 antibody, and Mr. Cornelius E. Regan for his technical support.
Footnotes
Financial interest: The authors declare no financial interests related to this work.
Author Contributions
S.L. and M.J. designed research; S.L., J.L., Y. Z., W.C.G., and M.J. performed research; J.M.H., N.G.B., and J.H.M. contributed unpublished reagents/analytic tools; S.L. and M.J. analyzed data; W.C.G. and J.H.M. edited the paper; S.L. and M.J. wrote the paper.
References
- 1.Battelle BA, LaVail MM. Rhodopsin content and rod outer segment length in albino rat eyes: modification by dark adaptation. Exp Eye Res. 1978;26:487–497. doi: 10.1016/0014-4835(78)90134-3. [DOI] [PubMed] [Google Scholar]
- 2.Bygum A, Westermark P, Brandrup F. Ichthyosis prematurity syndrome: a well-defined congenital ichthyosis subtype. J Am Acad Dermatol. 2008;59:S71–S74. doi: 10.1016/j.jaad.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Moiseyev G, Takahashi Y, Ma JX. Impacts of two point mutations of RPE65 from Leber's congenital amaurosis on the stability, subcellular localization and isomerohydrolase activity of RPE65. FEBS Lett. 2006;580:4200–4204. doi: 10.1016/j.febslet.2006.06.078. [DOI] [PubMed] [Google Scholar]
- 4.Cortina MS, Gordon WC, Lukiw WJ, Bazan NG. Light-induced photoreceptor damage triggers DNA repair: differential fate of rods and cones. Adv Exp Med Biol. 2003;533:229–240. doi: 10.1007/978-1-4615-0067-4_29. [DOI] [PubMed] [Google Scholar]
- 5.Danciger M, Matthes MT, Yasamura D, Akhmedov NB, Rickabaugh T, Gentleman S, Redmond TM, La Vail MM, Farber DB. A QTL on distal chromosome 3 that influences the severity of light-induced damage to mouse photoreceptors. Mamm Genome. 2000;11:422–427. doi: 10.1007/s003350010081. [DOI] [PubMed] [Google Scholar]
- 6.Deigner PS, Law WC, Canada FJ, Rando RR. Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science. 1989;244:968–971. doi: 10.1126/science.2727688. [DOI] [PubMed] [Google Scholar]
- 7.Golczak M, Maeda A, Bereta G, Maeda T, Kiser PD, Hunzelmann S, von Lintig J, Blaner WS, Palczewski K. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. J Biol Chem. 2008;283:9543–9554. doi: 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm C, Wenzel A, Hafezi F, Yu S, Redmond TM, Reme CE. Protection of Rpe65-deficient mice identifies rhodopsin as a mediator of light-induced retinal degeneration. Nat Genet. 2000;25:63–66. doi: 10.1038/75614. [DOI] [PubMed] [Google Scholar]
- 9.Gu SM, Thompson DA, Srikumari CR, Lorenz B, Finckh U, Nicoletti A, Murthy KR, Rathmann M, Kumaramanickavel G, Denton MJ, Gal A. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat Genet. 1997;17:194–197. doi: 10.1038/ng1097-194. [DOI] [PubMed] [Google Scholar]
- 10.Guignard TJ, Jin M, Pequignot MO, Li S, Chassigneux Y, Chekroud K, Guillou L, Richard E, Hamel CP, Brabet P. FATP1 inhibits 11-cis retinol formation via interaction with the visual cycle retinoid isomerase RPE65 and lecithin:retinol acyltransferase. J Biol Chem. 2010;285:18759–18768. doi: 10.1074/jbc.M109.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall AM, Wiczer BM, Herrmann T, Stremmel W, Bernlohr DA. Enzymatic properties of purified murine fatty acid transport protein 4 and analysis of acyl-CoA synthetase activities in tissues from FATP4 null mice. J Biol Chem. 2005;280:11948–11954. doi: 10.1074/jbc.M412629200. [DOI] [PubMed] [Google Scholar]
- 12.Jin M, Sawamoto K, Ito M, Okano H. The interaction between the Drosophila secreted protein argos and the epidermal growth factor receptor inhibits dimerization of the receptor and binding of secreted spitz to the receptor. Mol Cell Biol. 2000;20:2098–2107. doi: 10.1128/mcb.20.6.2098-2107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin M, Yuan Q, Li S, Travis GH. Role of LRAT on the retinoid isomerase activity and membrane association of Rpe65. J Biol Chem. 2007;282:20915–20924. doi: 10.1074/jbc.M701432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin M, Li S, Nusinowitz S, Lloyd M, Hu J, Radu RA, Bok D, Travis GH. The role of interphotoreceptor retinoid-binding protein on the translocation of visual retinoids and function of cone photoreceptors. J Neurosci. 2009;29:1486–1495. doi: 10.1523/JNEUROSCI.3882-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaschula CH, Jin M, Desmond-Smith NS, Travis GH. Acyl CoA:retinol acyltransferase (ARAT) activity is present in bovine retinal pigment epithelium. Exp Eye Res. 2006;82:111–121. doi: 10.1016/j.exer.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Kim SR, Fishkin N, Kong J, Nakanishi K, Allikmets R, Sparrow JR. Rpe65 Leu450Met variant is associated with reduced levels of the retinal pigment epithelium lipofuscin fluorophores A2E and iso-A2E. Proc Natl Acad Sci U S A. 2004;101:11668–11672. doi: 10.1073/pnas.0403499101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc Natl Acad Sci U S A. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klar J, Schweiger M, Zimmerman R, Zechner R, Li H, Torma H, Vahlquist A, Bouadjar B, Dahl N, Fischer J. Mutations in the fatty acid transport protein 4 gene cause the ichthyosis prematurity syndrome. Am J Hum Genet. 2009;85:248–253. doi: 10.1016/j.ajhg.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knott EJ, Sheets KG, Zhou Y, Gordon WC, Bazan NG. Spatial correlation of mouse photoreceptor-RPE thickness between SD-OCT and histology. Exp Eye Res. 2011;92:155–260. doi: 10.1016/j.exer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes VS, Gibbs D, Libby RT, Aleman TS, Welch DL, Lillo C, Jacobson SG, Radu RA, Steel KP, Williams DS. The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum Mol Genet. 2011;20:2560–2570. doi: 10.1093/hmg/ddr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda A, Maeda T, Golczak M, Imanishi Y, Leahy P, Kubota R, Palczewski K. Effects of potent inhibitors of the retinoid cycle on visual function and photoreceptor protection from light damage in mice. Mol Pharmacol. 2006;70:1220–1229. doi: 10.1124/mol.106.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda A, Maeda T, Golczak M, Chou S, Desai A, Hoppel CL, Matsuyama S, Palczewski K. Involvement of all-trans-retinal in acute light-induced retinopathy of mice. J Biol Chem. 2009;284:15173–15183. doi: 10.1074/jbc.M900322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiti P, Gollapalli D, Rando RR. Specificity of binding of all-trans-retinyl ester to RPE65. Biochemistry. 2005;44:14463–14469. doi: 10.1021/bi0510779. [DOI] [PubMed] [Google Scholar]
- 25.Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, Eliaou C, Liu SY, Harris E, Redmond TM, Arnaud B, Claustres M, Hamel CP. Mutations in RPE65 cause Leber's congenital amaurosis. Nat Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 26.Mata NL, Radu RA, Clemmons RC, Travis GH. Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron. 2002;36:69–80. doi: 10.1016/s0896-6273(02)00912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milger K, Herrmann T, Becker C, Gotthardt D, Zickwolf J, Ehehalt R, Watkins PA, Stremmel W, Fullekrug J. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J Cell Sci. 2006;119:4678–4688. doi: 10.1242/jcs.03280. [DOI] [PubMed] [Google Scholar]
- 28.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc Natl Acad Sci U S A. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moulson CL, Martin DR, Lugus JJ, Schaffer JE, Lind AC, Miner JH. Cloning of wrinkle-free, a previously uncharacterized mouse mutation, reveals crucial roles for fatty acid transport protein 4 in skin and hair development. Proc Natl Acad Sci U S A. 2003;100:5274–5279. doi: 10.1073/pnas.0431186100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moulson CL, Lin MH, White JM, Newberry EP, Davidson NO, Miner JH. Keratinocyte-specific expression of fatty acid transport protein 4 rescues the wrinkle-free phenotype in Slc27a4/Fatp4 mutant mice. J Biol Chem. 2007;282:15912–15920. doi: 10.1074/jbc.M701779200. [DOI] [PubMed] [Google Scholar]
- 31.Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 32.Nusinowitz S, Nguyen L, Radu R, Kashani Z, Farber D, Danciger M. Electroretinographic evidence for altered phototransduction gain and slowed recovery from photobleaches in albino mice with a MET450 variant in RPE65. Exp Eye Res. 2003;77:627–638. doi: 10.1016/s0014-4835(03)00217-3. [DOI] [PubMed] [Google Scholar]
- 33.Pang JJ, Chang B, Hawes NL, Hurd RE, Davisson MT, Li J, Noorwez SM, Malhotra R, McDowell JH, Kaushal S, Hauswirth WW, Nusinowitz S, Thompson DA, Heckenlively JR. Retinal degeneration 12 (rd12): a new, spontaneously arising mouse model for human Leber congenital amaurosis (LCA) Mol Vis. 2005;11:152–162. [PubMed] [Google Scholar]
- 34.Philp AR, Jin M, Li S, Schindler EI, Iannaccone A, Lam BL, Weleber RG, Fishman GA, Jacobson SG, Mullins RF, Travis GH, Stone EM. Predicting the pathogenicity of RPE65 mutations. Hum Mutat. 2009;30:1183–1188. doi: 10.1002/humu.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci U S A. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 37.Saari JC, Bredberg DL. CoA- and non-CoA-dependent retinol esterification in retinal pigment epithelium. J Biol Chem. 1988;263:8084–8090. [PubMed] [Google Scholar]
- 38.Sagara H, Hirosawa K. Monoclonal antibodies which recognize endoplasmic reticulum in the retinal pigment epithelium. Exp Eye Res. 1991;53:765–771. doi: 10.1016/0014-4835(91)90112-r. [DOI] [PubMed] [Google Scholar]
- 39.Sieving PA, Chaudhry P, Kondo M, Provenzano M, Wu D, Carlson TJ, Bush RA, Thompson DA. Inhibition of the visual cycle in vivo by 13-cis retinoic acid protects from light damage and provides a mechanism for night blindness in isotretinoin therapy. Proc Natl Acad Sci U S A. 2001;98:1835–1840. doi: 10.1073/pnas.041606498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon A, Hellman U, Wernstedt C, Eriksson U. The retinal pigment epithelial-specific 11-cis retinol dehydrogenase belongs to the family of short chain alcohol dehydrogenases. J Biol Chem. 1995;270:1107–1112. [PubMed] [Google Scholar]
- 41.Simon A, Romert A, Gustafson AL, McCaffery JM, Eriksson U. Intracellular localization and membrane topology of 11-cis retinol dehydrogenase in the retinal pigment epithelium suggest a compartmentalized synthesis of 11-cis retinaldehyde. J Cell Sci. 1999;112(Pt 4):549–558. doi: 10.1242/jcs.112.4.549. [DOI] [PubMed] [Google Scholar]
- 42.Sobol M, Dahl N, Klar J. FATP4 missense and nonsense mutations cause similar features in Ichthyosis Prematurity Syndrome. BMC Res Notes. 2011;4:90. doi: 10.1186/1756-0500-4-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl A, Hirsch DJ, Gimeno RE, Punreddy S, Ge P, Watson N, Patel S, Kotler M, Raimondi A, Tartaglia LA, Lodish HF. Identification of the major intestinal fatty acid transport protein. Mol Cell. 1999;4:299–308. doi: 10.1016/s1097-2765(00)80332-9. [DOI] [PubMed] [Google Scholar]
- 44.Szel A, Rohlich P, Caffe AR, Juliusson B, Aguirre G, Van Veen T. Unique topographic separation of two spectral classes of cones in the mouse retina. J Comp Neurol. 1992;325:327–342. doi: 10.1002/cne.903250302. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi Y, Chen Y, Moiseyev G, Ma JX. Two point mutations of RPE65 from patients with retinal dystrophies decrease the stability of RPE65 protein and abolish its isomerohydrolase activity. J Biol Chem. 2006;281:21820–21826. doi: 10.1074/jbc.M603725200. [DOI] [PubMed] [Google Scholar]
- 46.Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol. 2000;149:707–718. doi: 10.1083/jcb.149.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wenzel A, Reme CE, Williams TP, Hafezi F, Grimm C. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J Neurosci. 2001;21:53–58. doi: 10.1523/JNEUROSCI.21-01-00053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winston A, Rando RR. Regulation of isomerohydrolase activity in the visual cycle. Biochemistry. 1998;37:2044–2050. doi: 10.1021/bi971908d. [DOI] [PubMed] [Google Scholar]
- 49.Xue L, Gollapalli DR, Maiti P, Jahng WJ, Rando RR. A palmitoylation switch mechanism in the regulation of the visual cycle. Cell. 2004;117:761–771. doi: 10.1016/j.cell.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Yuan Q, Kaylor JJ, Miu A, Bassilian S, Whitelegge JP, Travis GH. Rpe65 isomerase associates with membranes through an electrostatic interaction with acidic phospholipid headgroups. J Biol Chem. 2010;285:988–999. doi: 10.1074/jbc.M109.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Sheets KG, Knott EJ, Regan CE, Jr, Tuo J, Chan CC, Gordon WC, Bazan NG. Cellular and 3D optical coherence tomography assessment during the initiation and progression of retinal degeneration in the Ccl2/Cx3cr1-deficient mouse. Exp Eye Res. 2011;93:636–648. doi: 10.1016/j.exer.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]