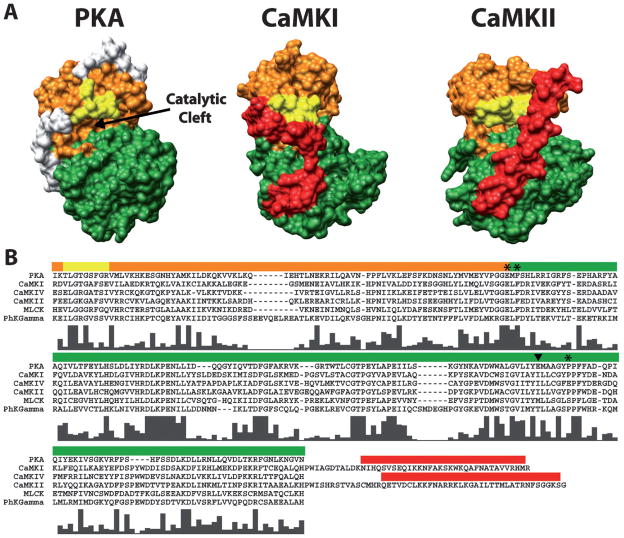
Figure 2.
Conservation of bi-lobed structure and functional regions within the catalytic domains of PKA, CaMKI and CaMKII. (A) Depicted is a surface rendering of crystal structures for PKA (PDB# 1J3H), CaMKI (PDB# 1A06) and CaMKII (PDB# 2BDW) catalytic domains. Conserved functional regions are colored orange (ATP-binding lobe), green (substrate-binding lobe) and yellow (glycine-rich loop). The autoinhibitory domains of CaMKI and CaMKII are colored red. The white regions on PKA are N- and C-terminal residues not conserved in the CaM-kinases. Note the conserved bi-lobed structure with a catalytic cleft at the interface between the lobes. (B) Multi-sequence alignment of amino acids spanning the catalytic domains of PKA and all the CaM-kinases (except CaMKIII due to its lack of homology). Colored bars above the sequences correspond to the amino acids which make up the functional regions of the structures above displayed in the same color. The residues C-terminal to the catalytic domains of CaMKI and CaMKII are shown in order to include their autoinhibitory domains. Histograms below the aligned residues show the high level of conserved amino acids in the catalytic domains of all the CaM-kinases with respect to each other and PKA. Amino acid positions marked by the asterisks (*) and the arrowhead correspond to the four amino acids in CaMKI involved in substrate binding. The amino acid marked by the arrowhead is of interest as it is conserved within the multifunctional but not the substrate-specific CaM-kinases. Sequences were aligned using ClustalX (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/).