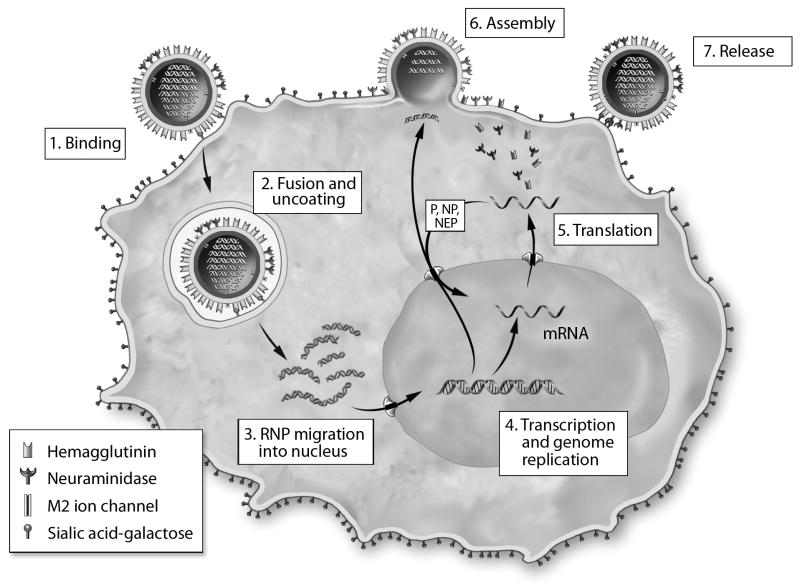
Figure 1.
The influenza A virus replication cycle. The virion core contains 8 RNA genome segments encapsidated by nucleoprotein (RNPs) and embedded together with associated polymerase (P) molecules in a matrix of M1 proteins. After binding to sialic acid-galactose linked to a cell-surface glycoprotein or glycolipid, the virion is taken up in an endocytic vesicle, where acidification activates cellular proteases that cleave the HA, leading to the release of a fusion peptide and a conformational change that bring viral and endosomal membranes together. Acidification also produces a flow of protons through the M2 ion channel into the interior of the virion, causing the RNPs to dissociate from the M1 matrix and be released into the cytoplasm. They are then transported to the nucleus, where a viral polymerase complex performs transcription and genome replication. The resulting mRNAs move to the cytoplasm and are translated, producing new RNP protein components that are transported back to the nucleus to associate with nascent genome segments. The exit of new RNPs from the nucleus is aided by the viral NS2 (nuclear export protein, NEP). Meanwhile, nascent HA, NA and M2 molecules pass through the Golgi apparatus and undergo glycosylation before moving to the cell membrane. Virion assembly occurs as RNPs and M1 proteins associate with cytoplasmic tails of HA and NA. Successful release of new virus particles requires that NA cleave sialic acid from galactose on the cell surface or on adjacent virions to prevent HA binding.