Fig. 5. Arm-type site and minimal DNA requirements in excisive recombination.
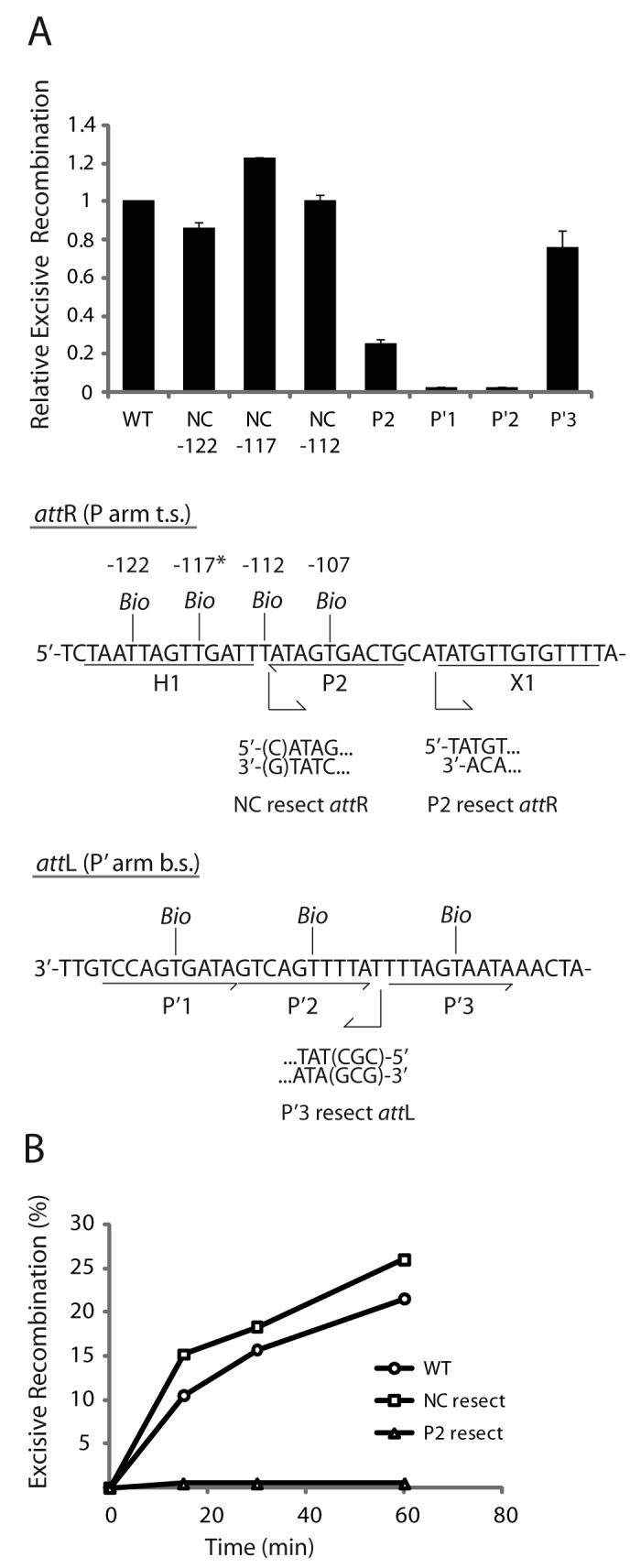
A. Biotin Interference Assay of excisive recombination. Att sites containing a single biotin dT substitution at one of the indicated positions were recombined with unsubstituted partners and the recombination efficiency was normalized to that of a control reaction with no substitutions (20-35% recombination efficiency). Based on the X-ray crystal structure (12), the biotin at −117 (*) is predicted to interfere with the putative non-canonical Int binding in the same way that biotin at position −107 blocks Int binding at P2. Biotin dT substitutions at positions −122 and −112 required changing the wild-type sequence from a G and a C, respectively; in each case the unsubstituted control attR was also mutated to T. Numbering of the att bases corresponds to the sequence of attP (14,52). B. Resection analysis of excisive recombination. Full-length attR substrate (WT) and substrates with the putative non-canonical site resected (NC resect) and P2 resected (P2 resect) were recombined with an attL lacking P′3 (P′3 resect), which is not required for excisive recombination (see text). Bent arrows beneath the attR and attL sequences in (A) denote resection endpoints and the direction of remaining sequence in the attR and attL substrates. The P2 resect substrate was generated by an Nde I digest and the P′3 substrate contained a CGC insertion after base +74 to stabilize the resected end. WT attR substrate contained a mutation in the H1 site to correct for the absence of the H1 site in the NC resect and P2 resect substrates. IHF binding at the H1 site inhibits excisive recombination and thus mutating or removing this site is stimulatory (16).
