Fig. 7. Formation of ternary HJ-tetrameric Int complexes with arm oligonucleotides encoding different combinations of P′ sites.
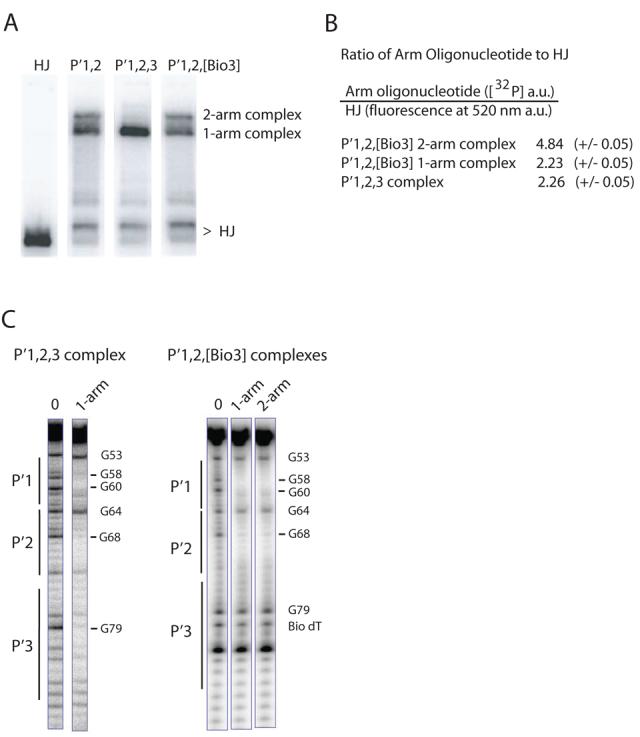
A. HJ-tetrameric Int-arm-type oligonucleotide ternary complexes were formed by incubating catalytically-inactive IntF (Y342F mutation), [32P]-radiolabeled Holliday junction, and a 40 bp oligonucleotide containing either the indicated unsubstituted or biotin dT-substituted arm-type P′ Int binding sites. The HJ lane lacks protein and arm oligonucleotide. Complexes were electrophoresed on a 7% polyacrylamide gel and visualized by autoradiography. B. Quantitation of the ratio of arm oligonucleotides to HJ in the P′1,2,3 and P′1,2,[Bio3] ternary complexes. [32P] signal intensity of P′1,2,3 and P′1,2,[Bio3] oligonucleotides and fluorescence intensity of a bodipy-FL labeled HJ were measured as described in Experimental Procedures and are reported in arbitrary units (a.u.). C. DMS protection of P′1,2,3 and P′1,2,[Bio3] 50 bp oligonucleotides (5′ [32P]-labeled on the bottom strand) complexed with HJ-tetrameric Int. Following complex formation, the reactions were treated with DMS, quenched, and electrophoresed on a 7% polyacrylamide gel. The indicated ternary complexes were excised from the gel, chemically cleaved at the methylated bases, and analyzed on a sequencing gel. Free P′1,2,3 and P′1,2,[Bio3] oligonucleotides served as the minus protein samples (0 lanes) to identify diminished (−) and enhanced (+) DNA cleavage sites. Numbering of the bases refers to the bottom-strand sequence of attP (52).
