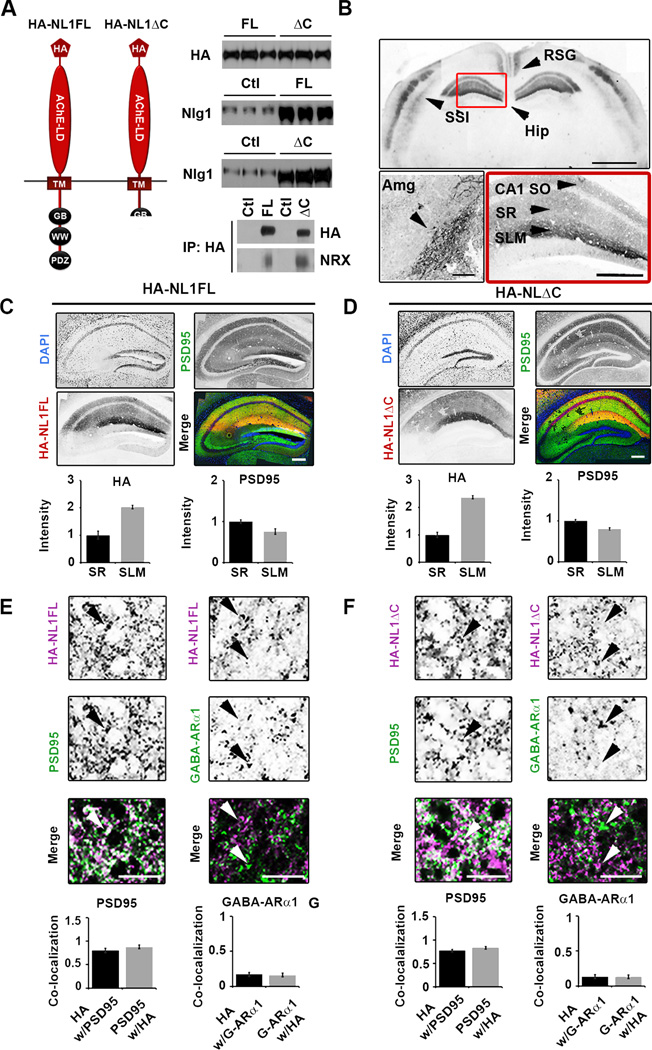
Figure 1. Expression of HA-NL1FL and HA-NL1ΔC in transgenic mice.
(A) Left, schematic of the protein structure of HA-NL1FL and HA-NL1ΔC encoded by the constructs that were used to generate transgenic mice. Abbreviations: Hemaglutinin epitope tag (HA), transmembrane domain (TM), gephyrin binding motif (GB), WW binding domain (WW), PDZ type II binding domain (PDZ). Right, absolute levels of HA compared between HA-NL1FL mice and HA-NL1ΔC mice (top panel), NL1 levels compared between transgenics and their controls (middle panels). 5ug total protein loaded into each lane. Immunoprecipitation using antibodies to HA and immunoblotting for Neurexins 1, 2 and 3 reveals similar level of interaction with HA-NL1FL and HA-NL1ΔC (p = 0.37, n = 2 independent IPs). (B) Immuno-labeling of HA shows localization patterns of the HA-NL1 constructs. Top panel: retrosplenial angular gyrus (RSG), layers II/III and V of somatosensory cortex (SS1) and the hippocampus (Hip), arrow heads, scale bar equals 2 mm. Lower left panel: Expression in the lateral amygdala (Amg), arrow head, scale bar equals 300 µm. Lower right panel: Expression in specific strata of the hippocampus, arrow heads, stratum oriens (SO), stratum radiatum (SR), stratum lacunosum moleculare (SLM). Scale bar equals 200µm. Dark areas reflect positive labeling. Sections analyzed were between −1.58 mm and −2.30 mm of bregma. (C & D) Immunolabeling of PSD95 (green), DAPI (blue) and HA epitope tag (red) for HA-NL1FL (C) and HA-NL1ΔC (D), scale bar equals 200µm. Quantification of both HA and PSD95 intensity levels between SLM and SR. All intensities normalized to SR levels. (E & F) Higher magnification and quantification of co-localization between HA and markers of excitatory (PSD95) vs. inhibitory (GABA-ARα1) synaptic markers, (E) HA-NL1FL mice and (F) HA-NL1ΔC mice. Differences in co-localization patterns in all cases were not significant. All data shown are mean ± SEM, significance determined by Student’s t-test, n = 3 paired littermates per test (3 double positives compared to 3 littermates of mixed single transgenic backgrounds).