Abstract
Purpose
Metastasis, the main cause of death from cancer, remains poorly understood at the molecular level.
Experimental design
Based on a pattern of reduced expression in human prostate cancer tissues and tumor cell lines, a candidate suppressor gene (SPARCL1) was identified. We used in vitro approaches to determine whether overexpression of SPARCL1 affects cell growth, migration, and invasiveness. We then employed xenograft mouse models to analyze the impact of SPARCL1 on prostate cancer cell growth and metastasis in vivo.
Results
SPARCL1 expression did not inhibit tumor cell proliferation in vitro. By contrast, SPARCL1 did suppress tumor cell migration and invasiveness in vitro and tumor metastatic growth in vivo, conferring improved survival in xenograft mouse models.
Conclusions
We present the first in vivo data suggesting that SPARCL1 suppresses metastasis of prostate cancer.
Keywords: prostate cancer, gene expression signature, meta-analysis, metastasis, SPARCL1 function in vivo
1. Introduction
In men, cancer of the prostate gland (CaP) is the most commonly diagnosed non-cutaneous malignancy, accounting for 29% of all cancer cases and the second most common cause of death by cancer in the USA. In 2012, an estimated 241,740 men were diagnosed with CaP and 28,170 men died of CaP (Siegel et al., 2012;Jemal et al., 2010). The majority of cancer-associated deaths and essentially all CaP deaths are due to metastases rather than primary tumor burden (Gupta and Massague, 2006). Thus, decreasing mortality of CaP depends on understanding the biology that underlies metastasis such as identification of genes involved in cancer metastasis that would benefit the design of more effective clinical intervention strategies. There is a wealth of evidence indicating that the acquisition of malignant progression and aggressive traits of cancer can be promoted or inhibited by a set of functional genes known as metastasis-regulatory genes in various cancers (Cher et al., 1999). These can be broadly categorized as pro-metastasis or metastasis-suppressor genes. Pro-metastasis genes drive conversion from non-metastatic to metastatic cells (Seraj et al., 2000). Metastasis-suppressor genes suppress the formation of metastases without affecting primary tumor growth (Kauffman et al., 2003), a characteristic that distinguishes them from tumor-suppressor genes.
To identify candidate metastasis-regulatory genes in CaP, a common and straightforward method is to identify a list of differentially expressed genes (expression signature) from analysis of transcriptional profiles of CaP correlated with poor prognosis. However, the single study-based signature is often underpowered, truncated, and low quality. These limitations can be overcome by combining related but independent studies into a meta-analysis for larger sample size and lower false discovery rate. There are a limited number of published CaP gene-expression studies having clinical survival outcome data for meta-analysis. We previously used a robust meta-analysis of gene expression profiles from hundreds of breast cancer datasets (Yi et al., 2007;Wu et al., 2009;Qiu et al., 2013). Using this approach, we discovered a novel and conserved gene expression signature predictive of metastasis risk in multiple cancers (breast, lung, and prostate cancer) (Qiu et al., 2013). We hypothesized that this expression signature is enriched for genes that are mechanistically involved with cancer metastasis including CaP. We tested this idea for a candidate gene, secreted protein acidic and rich in cysteine-like 1 (SPARCL1).
There are sporadic data illustrating down-regulation of SPARCL1 in lung (Bendik et al., 1998), colorectal (Yu et al., 2011), urinary bladder (Zaravinos et al., 2011), pancreatic (Esposito et al., 2007), and prostate cancers (Taylor et al., 2010;Chandran et al., 2007;Yu et al., 2004;Dhanasekaran et al., 2001;Bendik et al., 1998;Nelson et al., 1998;Hurley et al., 2012). Recombinant SPARCL1 inhibited spreading and adhesion of bovine aortic endothelial cells (Brekken et al., 2004) and endothelial cells on fibronectin substrates in vitro (Girard and Springer, 1996). When its function was assessed using cancer cell lines, SPARCL1 inhibited pancreatic (Esposito et al., 2007) and prostate cancer cell migration and invasion in vitro but did not restrict the growth of prostate cancer cells (Hurley et al., 2012), suggesting that SPARCL1 is a potential suppressor of metastatic progression in prostate cancer. However, all previous results on SPARCL1 in CaP were derived from in vitro studies and clinical correlations. No in vivo data have been published to determine whether SPARCL1 contributes to CaP metastasis. Experiments, using a colon cancer cell line overexpressing SPARCL1 and a complementary model, suggested that SPARCL1 could reduce cell proliferation, anchorage-independent growth, and invasion in vitro and significantly inhibited orthotopic tumor growth in vivo. On this basis, Hu et al concluded that SPARCL1 functions as a tumor suppressor in colon cancer (Hu et al., 2012). The question of whether SPARCL1 can suppress metastasis in CaP in vivo has not previously been addressed.
Consistent with previous studies, we found that SPARCL1 was down regulated among human prostate tissue specimens and cell lines representing various levels of tumorigenicity and metastatic tendencies. However, we show here, in a prostate cancer model, that SPARCL1 does not inhibit tumor cell growth in vitro but does suppress tumor metastasis in vivo. Overexpression of SPARCL1 decreased the metastatic potential of human CaP (PC3) cells in both in vitro functional assays and in vivo experimental metastasis models. Specifically, SPARCL1 expression significantly inhibited tumor cell invasiveness and migration in vitro and capacity to metastasize to distant organs in vivo. These observations suggest that SPARCL1 can suppress metastasis in human CaP.
2. Materials and methods
2.1. Meta-analysis of human cancer profiles
The methods used for signature extraction, signature database development, and EXALT analysis were previously reported (Yi et al., 2007;Wu et al., 2009). Iterative EXALT analysis for identification of the 50-gene expression signature and its association with CaP metastasis has been described elsewhere (Qiu et al., 2013).
2.2. Clinical data
Clinical and gene-expression data for the 50-gene signature validation were obtained from independently published human cancer studies and the Gene Expression Omnibus (GEO) provided by the National Center for Biotechnology Information (NCBI).
2.3. Cell lines and primary tumor specimens
Cell lines were kindly provided as follows: PC3 and LNCaP human prostate carcinoma cell lines from ATCC (American Type Culture Collection, Manassas, VA, USA); the bioluminescent human prostate carcinoma cell line (PC3-Luc) from Dr. K. Pienta (University of Michigan Medical Center); non-tumorigenic human prostate epithelial cell lines NHPrE1 and BHPrE1 from our own stocks (Jiang et al., 2010); ARCaPM cells were from Novicure Biotechnology (Birmingham, AL, USA). De-identified human malignant and nonmalignant prostate tissue samples were collected and frozen immediately after surgical resection through the Vanderbilt Cooperative Human Tissue Network via the Department of Pathology in accordance with Vanderbilt IRB protocols.
PC3, PC3-Luc, and LNCaP cells were cultured at 37°C 5% CO2 in RPMI1640 containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. ARCaPM cells were cultured in MCaP-medium supplied by Novicure. Both NHPrE1 and BHPrE1 cells were cultured in HPrE-conditional medium described previously (Jiang et al., 2010).
2.4. Western Blotting
Equal amounts of cell or tissue lysates per lane were loaded onto 10% SDS polyacrylamide gels. Membranes were incubated with chicken polyclonal antibody specific for human SPARCL1 (Abcam, Cambridge, United Kingdom) at a 1:2000 dilution, and then the membranes were re-probed using mouse monoclonal antibody specific for human anti-β-actin (Sigma Chemical Company, Saint Louis, MO, USA).
2.5. Construction of a SPARCL1 expression vector and establishment of stable PC3-Luc cells overexpressing SPARCL1
The human gene SPARCL1 ORF (RC207583, OriGene Technologies, Inc, Rockville, MD) was subcloned into pBMN-I-GFP (Addgene Inc., Cambridge, MA, USA) to obtain a pBMN-SPARCL1-I-GFP plasmid. DNA sequencing was performed to verify the sequence of the constructed plasmid.
Plasmids pBMN-SPARCL1-I-GFP and pBMN-I-GFP were transfected into Phoenix cells by FuGENE 6 (Roche Applied Science, Indianapolis, IN, USA). Retroviral particles were harvested and used to infect PC3-Luc cells. GFP-positive PC3-Luc cells, either overexpressing SPARCL1 (PC3-luc/SPARCL1) or empty control (PC3-luc/EV) were collected by fluorescence-activated cell sorting.
2.6. Cell proliferation assay
PC3-luc/EV, PC3-luc/SPARCL1, PC3, and ARCaPM cells were plated at a density of 10,000 cells/well in Costar 96-well cell culture plates (CORNING, Tewksbury, MA, USA), respectively. PC3 and ARCaPM were treated with and without recombinant human SPARCL1 (hSPARCL1) (10 μg/ml). After incubation at various time points (day 1, day 2, and day 3), 20 μl MTT [3-(4, 5-methylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide] solution (5 mg/ml; Sigma) per well was added for 1 hour. Colorimetric changes were read on a microtiter plate reader with a 570-nm filter. Cell viability was estimated by a standard MTT assay (Price and McMillan, 1990) at various time intervals.
2.7. Colony formation in soft agarose
PC3-luc/EV, PC3-luc/SPARCL1, PC3, and ARCaPM cells (1 × 104) were suspended in complete medium with a top layer of 0.3% agarose and a bottom layer of 0.6% agarose in triplicate in 6-well plates. Among them, PC3 and ARCaPM cells were treated with and without recombinant human SPARCL1 (hSPARCL1) (10 μg/ml). Complete medium was changed every week. Colony formation was examined after 3 weeks. The colonies were stained with 500 μl MTT solution (5mg/ml; Sigma)/well for 1 h. Scanned images of the colonies were analyzed with the Image software.
2.8. Wound healing assay
A 12-well culture plate was placed on the magnetic platform after which each well was set up with magnetically attachable stencils (MAts) (Ashby et al., 2012) PC3-luc/EV or PC3-luc/SPARCL1 (350,000 cells/well) were added to each well and allowed to adhere. After overnight incubation, MAts were removed leaving behind wounds with even-edges and equal-distances. Images were taken immediately upon MAts removal at 0h and at 12h. Percent wound closure was quantified using T-scratch software (http://www.cse-lab.ethz.ch/).
2.9. Transwell migration assay
To assay cell migration, PC3 and ARCaPM were treated with recombinant human SPARCL1 (hSPARCL1) (10 μg/ml) (R&D Systems Inc., Minneapolis, MN). Cell migration was assayed using the Cell Migration Colorimetric Assay Kit (Millipore) according to the manufacturer’s instructions.
2.10. Transwell invasion assay
A cell invasion assay was performed using 6-well Transwell polycarbonate membrane inserts with 8.0-μm pores (CoStar) coated from the bottom with Matrigel (BD Biosciences, San Jose, CA, USA) as an extracellular matrix barrier. PC3-luc/EV or PC3-luc/SPARCL1 or ARCaPM cells (1 × 104) were detached and seeded in the upper chamber and cultured in serum-free medium for 48 h. ARCaPM cells were treated either with or without recombinant human SPARCL1 (hSPARCL1) (10 μg/ml) in the upper chamber medium. Cells were allowed to migrate towards medium containing 10% FBS in the bottom chamber. The non-migratory cells on the upper membrane surface were removed with a cotton tip, and the invasive cells attached to the lower membrane surface were fixed with 10% neutral buffered formalin (Thermo Fisher Scientific, Waltham MA, USA) and stained with modified Mayer’s hematoxylin (Thermo Fisher Scientific). The numbers of migrated cells were counted in 3 randomly selected fields under a microscope. Data presented are representative of three individual wells and repeated twice.
2.11. In vivo animal experiment studies
Severe combined immunodeficient (SCID) male mice (Harlan, Indianapolis, IN) were housed and maintained under specific pathogen-free conditions in facilities approved by the Vanderbilt University Institutional Animal Care and Use Committee. Mice were kept at least 1 week before experimental manipulation.
For orthotopic xenografting (OX) experiments, 10-week-old SCID mice were randomized into two groups: PC3-luc/EV and PC3-luc/SPARCL1. PC3-luc/EV or PC3-luc/SPARCL1 cells (3×105 in 30 μl) mixing with neutralized collagen gel were implanted into the mouse anterior prostate (AP) lobe through a lower midline laparotomy incision (Park et al., 2010). The liquid collagen gel containing PC3 cells became solid in vivo. We then closed the incision of AP to let the solid gel piece stay inside of the lumen of the AP lobe. There was no cell suspension left in the space surrounding space of the prostatic ducts. After PC3 xenografting, mice were imaged biweekly for bioluminescence using an in vivo Imaging System (IVIS) to monitor tumor growth. The end point for overall survival time of each animal was recorded and determined using a panel of clinical parameters including muscle wasting, loss of fat deposits, and prominent bones (Yang et al., 2011). Animals were weighed at the initiation of the experiment and monitored weekly. When the predetermined clinical parameters were detected, the animals were weighed daily. If more than 20% of their starting body weight was lost, mice were euthanized. Upon sacrifice, the prostate and other organs were removed for imaging and histological examination. Survival data were plotted on a Kaplan-Meier curve, and the two groups were compared using the Log-rank (Mantel-Cox) test (the open-source R software, version 2.14.1 at www.r-project.org).
Intracardiac (IC) injections were performed as previously described (Park et al., 2010). In brief, 6–8-week-old SCID male mice were randomized into two groups. PC3-luc/EV or PC3-luc/SPARCL1 cells (5 × 105) were re-suspended in 100 μl PBS and slowly injected into the left ventricle of the mice. IVIS and radiographs (Faxitron) were used to confirm successful injections into the mouse body and to monitor metastasis formation.
2.12. Bioluminescence imaging
Mice bearing PC3-luc/EV or PC3-luc/SPARCL1 tumors were imaged for bioluminescent signal on a weekly basis as previous described (Yang et al., 2011). Mice were anesthetized and imaged using an IVIS Imaging system 200 (Xenogen Corp., Alameda, CA). Tumor burden was measured based on total photons per second with background subtraction per region of interest (ROI).
Following ex vivo bioluminescence imaging, selected organs were fixed in 10% neutral buffered formalin. Tissues were then processed, embedded in paraffin, sectioned at 4 μm, and stained with hematoxylin and eosin (H.E.).
3. Results
3.1. Discovery of candidate suppressor genes
We recently reported an association between a 50-gene expression signature and CaP metastasis (Qiu et al., 2013). We hypothesized that genes enriched in the 50-gene signature are mechanistically involved in CaP metastasis. In metastatic tumors, the changes in expression of pro-metastasis and metastasis-suppressor genes are expected to be divergent (pro-metastasis genes being up-regulated while suppressors are down-regulated) (Weigelt et al., 2005). Eleven of the 50 genes were downregulated in aggressive tumors. We used a bioinformatic approach to determine if any of the 11 suppressor candidate genes exhibited downregulation in human CaP (Wu et al., 2009). We surveyed gene expression of the 11 candidates among data from 10 published transcriptional profiling studies performed on normal or diseased prostate tissues (Fig. 1).
Figure 1. Co-expression analysis of the 11 suppressor candidates from the 50-gene expression signature among CaP studies.
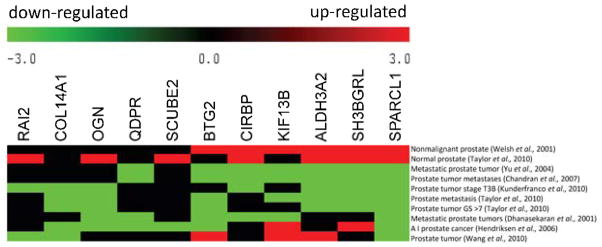
Meta-heat map depicts 11 suppressor candidate gene expression patterns across 10 transcriptional profiling studies in normal or diseased prostate tissue. The different expression profiling studies are represented in rows, and the 11 candidate genes are represented in columns. The colors in the heat map represent the direction of differential gene expression within a given transcriptional profile (red for up, green for down, and black for a missing match). Color intensity reflects the confidence levels of differential expression.
Differential gene expression (up or down) was determined by a comparison between an experimental and control group within each dataset, and the relative change of expression in candidate genes was represented in rows by an experimental sample group. Out of the 11 candidate genes, SPARC-like 1 (SPARCL1) displayed the most consistent profile among the 10 datasets (Fig. 1). Upregulation of SPARCL1 was found in nonmalignant (benign) (Welsh et al., 2001) and normal prostate tissues (Taylor et al., 2010). Downregulation of SPARCL1 was observed in CaP samples (Wang et al., 2010), tumors with high grade (T3B) (Kunderfranco et al., 2010), high Gleason scores (GS > 7) (Taylor et al., 2010), androgen independent (AI) status (Hendriksen et al., 2006), and metastatic prostate tumors (Taylor et al., 2010;Chandran et al., 2007;Yu et al., 2004;Dhanasekaran et al., 2001). Based on the ability of the related protein SPARC’s influence on tumor progression, invasion, and metastasis (Clark and Sage, 2008), we hypothesized that SPARCL1 can suppress metastasis in human CaP.
3.2. SPARCL1 protein expression is lost in invasive human CaP
To determine whether SPARCL1 protein expression is decreased in human CaP, we evaluated SPARCL1 expression by Western blot in protein lysates from five human patients with high-grade CaP and one non-malignant tissue (Fig. 2A). Relative to a loading control β-actin, SPARCL1 levels were downregulated in CaP samples in comparison to the non-malignant sample. We examined SPARCL1 expression patterns in a panel of human prostate cell lines (Fig. 2B). SPARCL1 expression is undetectable in LNCaP, ARCaPM, and PC3 cancer cells. In contrast, high expression of SPARCL1 was observed in benign human prostate tissue and benign prostate cell lines (NHPrE1) (Fig. 2B).
Figure 2. SPARCL1 expression profiles in human prostate.
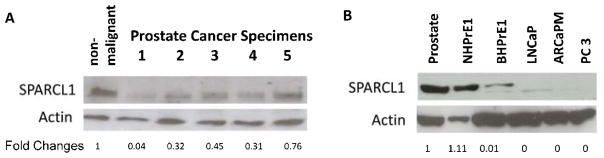
(A) Prostate protein samples were derived from benign human prostate tissue and high-grade CaP specimens. SPARCL1 expression was measured using Western Blot (upper panel), and total protein amounts were examined by re-probing with anti-β-actin (middle panel). Western blot results were quantified by densitometry. Normalized fold changes of SPARCL1 expression between each CaP sample and non-malignant sample were deduced (fold change values) and listed beneath the SPARCL1 bands (bottom panel). Note that CaP samples expressed lower levels of endogenous SPARCL1 than non-malignant prostate samples.
(B) Evaluations of SPARCL1 expression profiles in human CaP cell lines. A Western blot was used to measure differential expression of SPARCL1 among several CaP cell lines. Cell lysate samples included benign human prostate tissue and cell lines (NHPrE1/BHPrE1) and various CaP cell lines (LNCaP, ARCaPM, and PC3). The Western blot membrane was re-probed with anti-β-actin. The SPARCL1 fold changes were derived from the ratios to β-actin expression within each cell line and were further normalized by the ratio from the normal prostate sample.
3.3. Ectopic SPARCL1 expression decreases in vitro metastatic potential
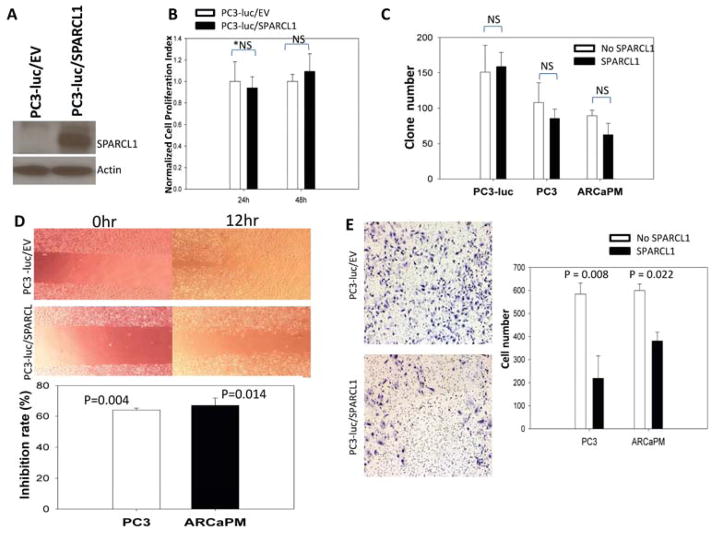
To determine the impact of restoring SPARCL1 expression in a model of aggressive human CaP, we ectopically expressed SPARCL1 or an empty vector control (EV) in luciferase-expressing PC3 cells (PC3-luc), resulting in PC3-luc/SPARCL1 and PC3-luc/EV cells, respectively. The SPARCL1 expression was confirmed in PC3-luc/SPARCL1 cells (Fig. 3A). PC3-luc/EV and PC3-luc/SPARCL1 cells were used to determine the effects of SPARCL1 expression on tumorigenicity and metastasis. A series of in vitro functional assays were performed (Fig. 3B–E).
Figure 3. in vitro evaluation of SPARCL1 function.
(A) Western Blot was used to confirm SPARCL1 expression in PC3-luc/SPARCL1 in comparison to control PC3-luc/EV cells. The Western blot membrane was re-probed with anti-β-actin.
(B) PC3-luc/EV control and PC3-luc/SPARCL1 cells were seeded into quadruplicate wells of 96-well plates and their proliferation rates determined by MTT assay. Data were plotted as mean relative growth+/−SEM.
(C) Anchorage-independent growth of CaP cells with SPARCL1 in soft agarose. PC3 and ARCaPM cells were treated with and without recombinant SPARCL1 (10 μg/ml). PC3-luc/EV, PC3-luc/SPARCL1, PC3, and ARCaPM cells were examined for their anchorage-independent growth in soft agarose. The comparisons of colony formation are plotted as a bar graph.
(D) Motility and migration analysis by wound healing assay (upper panel) and transwell migration assay (bottom panel). Upper panel, pictures were taken at 0 h and at 12 h. Wound healing closure percentages were compared between PC3-luc/EV and PC3-luc/SPARCL1. Bottom panel, a vertical bar graph depicts migration inhibition percentages in transwell assay from recombinant SPARCL1 treated PC3 and ARCaPM cells in comparison with corresponding untreated control cells.
(E) Images (left panel) and a bar graph of invasion cell counts (right panel) from representative transwell invasion assays. The invasion PC3 cells (PC3-luc/EV and PC3-luc/SPARCL1) and ARCaPM cells treated with recombinant SPARCL1 (X-axis in the bar graph) were examined for their invasiveness in transwell Matrigel invasion assays. The invasiveness is expressed in numbers of cell invading Matrigel in Y-axis. The statistics analysis used for panels (B, C, D, E) is Student’s t test. *NS: non-significant.
Proliferation rates of the PC3-luc/EV and PC3-luc/SPARCL1 cells were similar in 24- and 48-hour measurements (Fig. 3B), demonstrating that expression of SPARCL1 had no significant effect on PC3-luc cancer cell growth in vitro. Additional cell proliferation experiments were performed using PC3 and ARCaPM at multiple time points. The growth activities of both cell lines from day 1 to day 3 did not change significantly in the presence of recombinant SPARCL1 (Supplemental Fig. S1) in comparison to a corresponding control cell lines (P > 0.05). We then examined whether SPARCL1 could affect colony formation by plating the PC3-luc/EV, PC3-luc/SPARCL1, PC3, and ARCaPM cells in soft agarose. After 3 weeks of culture, the numbers of colonies formed were slightly lower when the CaP cells were cultured in the presence of recombinant SPARCL1. However, there was no significant difference of colony formation (P > 0.05) between control groups (PC3-luc/EV, PC3, and ARCaPM) and corresponding SPARCL1 treated groups, including PC3-luc/SPARCL1, PC3, and ARCaPM cells (Fig. 3C). The data indicate that SPARCL1 did not alter anchorage-independent growth of PC3 and ARCaPM cells.
Because SPARCL1 had no effect on cell proliferation or anchorage independent growth, we sought to determine whether overexpression of SPARCL1 changed cell motility and migration using an in vitro wound healing assay (Fig. 3D, upper panel). At 12 hours, PC3-luc/SPARCL1 cells showed significantly less motility and migration (wound-healing closure percentage, 15%; P = 3.2 × 10−7) when compared to the control (PC3-luc/EV, 33%). When CaP cells (PC3 and ARCaPM) were treated with or without recombinant SPARCL1 in a transwell migration assay, consistently, we found that recombinant SPARCL1 significantly inhibited both PC3 and ARCaPM cells across a transwell membrane in comparison with untreated control cells (inhibition rates, 64% and 67%, respectively; Fig. 3D, lower panel).
We also evaluated the SPARCL1 effect on invasive capability of PC3 cells (PC3-luc/EV and PC3-luc/SPARCL1) and ARCaPM cells. In a 48-hour transwell-invasion assay, more PC3-luc/EV cells invaded through the Matrigel coated basement membrane compared to PC3-luc/SPARCL1 cells (Fig. 3E, left panel). In comparison to the empty vector control (PC3-luc/EV) cells, SPARCL1 significantly decreased the invasiveness of PC3 cells by 2.7-fold in a transwell Matrigel invasion assay (Fig. 3E, right panel, P =0.008). We found that recombinant SPARCL1 could also reduce invasiveness of ARCaPM cells (Fig. 3E, right panel, P = 0.022).
Results from these in vitro functional experiments suggest that SPARCL1 expression does not alter PC3-luc growth under either anchorage-dependent or independent conditions. However, SPARCL1 suppressed prostate cancer cell migration and invasion.
3.4. Effect of SPARCL1 on prostate cancer metastasis
Because SPARCL1 could suppress motility and invasion in vitro, we evaluated the effect of SPARCL1 on the development of metastases in vivo. To test this, we introduced PC3-luc/EV and PC3-luc/SPARCL1 cells into a mouse prostate (orthotopic xenograft [OX] model), (Fig. 4) and mouse arterial circulation via intracardiac injection (IC model, Fig. 5) (Park et al., 2010).
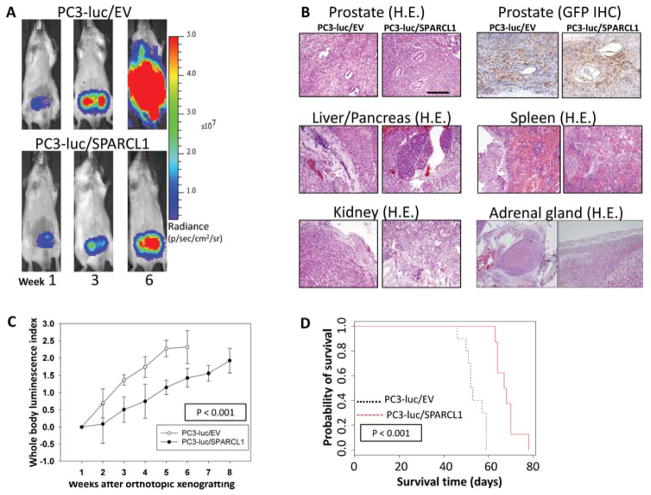
Figure 4. In vivo monitoring of orthotopic PC3 prostate tumor growth and metastasis with or without overexpression of SPARCL1.
(A) Representative ventral images of bioluminescence measurements for xenografted PC3 tumor growth and metastasis. Luciferase-tagged PC3-luc/EV and PC3-luc/SPARCL1 cells were surgically implanted into the anterior prostate of male SCID mice. Ventral images of anesthetized representative control (PC3-luc/EV) and experimental (PC3-luc/SPARCL1) mice are shown at weeks 1, 3, and 6 to monitor orthotopic prostate tumor growth and tumor spread to other organs. All images were set using the same pseudocolor scale in radiance defined as photons/second/cm2/steradian (p/sec/cm2/sr) to show relative bioluminescent changes over time.
(B) Histological slides include orthotopic tumors and metastatic tumors of PC3-luc/EV and PC3-luc/SPARCL1. Representative images were derived from mouse prostate stained with hematoxylin and eosin (H&E) (upper left panel). The same tissue sections were analyzed by IHC using an anti-GFP antibody (upper right panel) to identify human PC3 cells in both PC3-luc/EV and PC3-luc/SPARCL1 samples. The original xenografted GFP-positive PC3 cells were found in xenografts tumor site. Representative H&E-stained sections of metastasis organ specimens were derived from PC3-luc/EV and PC3-luc/SPARCL1 mice (middle and bottom panels). The metastatic tumors were dissected from liver, pancreas, adrenal gland, kidney, and spleen. Scale bar = 100 μm.
(C) PC3 growth in mice. Whole body luminescence index shown as a function of time is displayed on the Y-axis as a measure of tumor burden. Bioluminescent signals emitted from the PC3 tumors were quantified in photons/s/cm2 at each imaging time. Using week 1 time point as a base value, mean tumor bioluminescence for each group per time point was normalized as mean log ratios. The plotted curves over time were used to compare the control PC3-luc/EV mice (n = 10, open circles) with the PC3-luc/SPARCL1 mice (n = 8, closed circles). The PC3-luc/SPARCL1 cells grew less aggressively with a statistically significant difference (P < 0.001) based on ANOVA analysis.
(D) Kaplan-Meier analysis for overall survival. Total survival data were stratified into 2 groups (EV and SPARCL1 mouse groups) by SPARCL1 expression. The survival plot illustrates two types of overall survival, a poor prognosis group (black dashed line, EV mouse group) and a good prognosis group (red solid line, SPARCL1 mouse group). The overall survival time in days is displayed on the X-axis, and the Y-axis shows the probability of overall time survival. The end point for overall survival time of each animal was recorded and determined using a panel of clinical parameters including muscle wasting, loss of fat deposits, and prominent bones. The log-rank test p-value indicates the statistical significance of survival time differences between the two groups (P < 0.0001). Mice inoculated with PC3-luc/SPARCL1 cells had a better survival probability.
Figure 5.
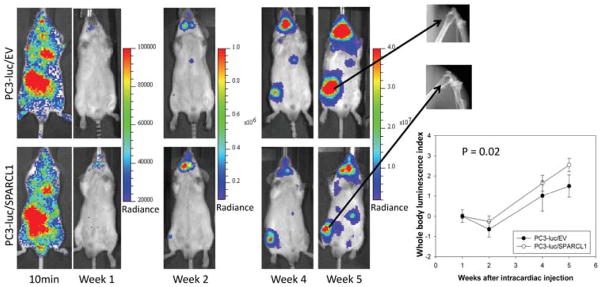
Representative ventral images of bioluminescence measurements for xenografting PC3 tumor metastasis. Luciferase-tagged PC3-luc/EV and PC3-luc/SPARCL1 cells were introduced into mouse arterial circulation via intra-cardiac injection (intracardiac model or IC model) of male SCID mice. Ventral images of anesthetized representative control (upper panel, PC3-luc/EV, 10 mice) and experimental (bottom panel, PC3-luc/SPARCL1, 8 mice) mice are shown at 10 minutes and week 1, 2, 4, and 5 for monitoring tumor spreading to organs and bones. At each time point, luciferase images were set using the same pseudocolor scale in radiance (photons/second/cm2/steradian) to show relative bioluminescent changes. The X-ray images (upper right panel) of metastatic PC3 tumors in femurs 5 weeks post-injection show osteolytic lesions in both control and experimental group mice. Whole body luminescence index shown as time series curves (lower right panel) is displayed on the Y-axis as a measure of total tumor burden. Bioluminescent signals emitted from the PC3 tumors were quantified in average values of radiance at each imaging time. Using week 1 time point as a base value, mean tumor bioluminescence for each group per time point was normalized as mean log ratios. The plotted curves over time were used to compare the control PC3-luc/EV mice (n = 10, open circles) with the PC3-luc/SPARCL1 mice (n = 8, closed circles). The difference (P = 0.02) between PC3-luc/EV mice and the PC3-luc/SPARCL1 mice was analyzed based on ANOVA analysis.
In either OX or IC mouse model, SCID mice xenografted with PC3 tumors were then monitored for the development of tumors at regular time intervals using noninvasive bioluminescence imaging. Representative bioluminescence images are presented from mice in OX model taken at 1, 3, and 6 weeks (Fig. 4A) or mice in IC model at 10 minutes and 1, 2, 4, and 5 weeks (Fig. 5) post-xenografting. Data measured as photon flux are shown in the same color scale for all mice between control mouse group (Fig. 4A upper panel and Fig. 5 upper panel, PC3-luc/EV) and SPARCL1 overexpression group (Fig. 4A lower panel and Fig. 5 lower panel, PC3-luc/SPARCL1). These results demonstrated significant differences in total tumor burden (Fig. 4A left panel and Fig. 5) and metastases in multiple distant organ locations including liver, spleen, pancreas, adrenal gland, and kidney, which were confirmed by dissected organ imaging (Supplemental Fig. S2). Osteolytic lesions were verified by X-ray images (Fig. 5 right upper panel). In both OX and IC models, mice implanted with PC3-luc/SPARCL1 cells displayed smaller areas and less intense luciferase activities in the whole body compared to the empty vector control mice (PC3-luc/EV), demonstrating that SPARCL1 expression could inhibit the development of metastases.
Histological analysis of OX PC3-luc/EV and PC3-luc/SPARCL1 tumors indicated that they had a similar pattern of invasive tumor cell growth in the prostate (Fig. 4B, upper panel) and other metastatic sites (Fig. 4B, middle and lower panel). To confirm this PC3 cell identity in xenografted tumors, IHC staining of GFP (PC3-luc cell marker, Fig. 4B, upper right panel) was performed to show that these tumors originated from xenografted PC3 cells (PC3-luc/EV and PC3-luc/SPARCL1). This observation confirmed primary PC3 tumor growth and metastasis in xenografing mouse models.
Quantification of whole-animal photon emission rates (Fig. 4C and Fig. 5 lower right panel) allowed us to gauge relative tumor burden in mice over time. We computed the whole body luminescence index from normalized and time-matched whole-body photon emission rates (reflective of total tumor burden, Log scale). From week 2 to 6 (Fig. 4C), the whole body luminescence indexes from PC3-luc/SPARCL1 group were consistently lower by approximately 4- to 7-fold than those from PC3-luc/EV group (P < 0.001). The total luminescence indices from week 1 to 5 in the PC3-luc/SPARCL1 group from IC model were also significantly lower than those in the PC3-luc/EV group (P = 0.02, Fig. 5, lower right panel). These results suggest that SPARCL1 inhibits the invasive growth of CaP and the progression of metastasis.
Kaplan-Meier curves were generated to evaluate overall survival for control mice (PC3-luc/EV) and mice xenografted with SPARCL1 overexpression cells (PC3-luc/SPARCL1). The results (Fig. 4D) indicated a significant difference in overall survival between the mice inoculated with PC3-luc/SPARCL1 and mice inoculated with PC3-luc/EV (P < 0.0001). Overall, mice inoculated with PC3-luc/SPARCL1 cells survived about two weeks longer. Mice bearing PC3-luc/SPARCL1 tumors had a better prognosis, as demonstrated by 100% survival at 60 days compared to 0% for mice harboring control cells (PC3-luc/EV). Thus, mice xenografted with PC3-luc/SPARCL1 had a decreased metastatic burden and longer overall survival.
3.5. SPARCL1 expression suppresses metastasis
To gain insight into whether SPARCL1 had an effect on the development and tropism of metastases, we evaluated the incidences of metastasis and tissue distribution of metastatic lesions in PC3-luc/EV and PC3-luc/SPARCL1 groups using both OX and IC models. The numbers of metastatic sites were determined using bioluminescence imaging of anesthetized mice, bioluminescence ex vivo of isolated organs (Fig. 4A), and X-ray imaging of the mouse skeletal system (Fig. 5).
Table 1 summarizes the results from both OX and IC models. OX model is a true test of metastasis while IC injection is a model that determines the ability of a cancer cell to successfully disseminate and colonize distal organs such as bone. In both models, PC3-luc/EV and PC3-luc/SPARCL1 cells were able to grow as visceral metastases in a variety of soft tissue sites such as liver, pancreas, adrenal gland, kidney, and spleen with a similar propensity and pattern. Using the IC model, both PC3-luc/EV and PC3-luc/SPARCL1 could spread to the bone with osteolytic responses at week 5 post IC injections (Fig. 5). Review of metastasis from sacrificed mice in both models indicated that frequency of metastases for both soft tissues and bones are consistently lower in PC3-luc/SPARCL1 mice than those in PC3-luc/EV mice (Table 1). When compared to PC3-luc-EV group, the total numbers of final visceral metastasis for the PC3-luc/SPARCL1 group were statistically significantly lower in both in vivo models, more specifically, 27% lower in the OX model (P = 0.01) and 45% lower in the IC model (P < 0.001). In the IC model, metastatic lesions among skeletal sites decreased in PC3-luc/SPARCL1 group but the difference was not statistically significant between PC3-luc/EV and PC3-luc/SPARCL1 (P = 0.07).
Table 1.
Frequency of metastases at various anatomical sites
| Models | Orthotopic | Intra-cardiac | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| PC3 cells | EV | SPARCL1 | P value | EV | SPARCL1 | P value |
| Visceral sites | ||||||
| Liver | 80(8/10)* | 63(5/8) | 90(9/10) | 75(6/8) | ||
| Pancreases | 60(6/10) | 25(2/8) | 67(6/9) | 25(2/8) | ||
| Spleen | 38(3/8) | 38(3/8) | 44(4/9) | 13(1/8) | ||
| Adrenal glands | 90(9/10) | 50(4/8) | 80(8/10) | 13(1/8) | ||
| Kidney | 100(10/10) | 63(5/8) | 80(8/10) | 13(1/8) | ||
| Total metastasis | 75(36/48) | 48(19/40) | 0.01 | 73(35/48) | 28(11/40) | < 0.001 |
| Skeletal sites | ||||||
| Tibia | NA** | NA | 40(8/20) | 13(2/16) | ||
| Femur | NA | NA | 30(6/20) | 19(3/16) | ||
| Mandible | NA | NA | 70(7/10) | 38(3/8) | ||
| Iliac crest | NA | NA | 5(1/20) | 6(1/16) | ||
| Ankle/paw | NA | NA | 3(1/40) | 0(0/32) | ||
| Humerus shoulder | NA | NA | 15(3/20) | 13(2/16) | ||
| Total metastases | NA | NA | 20(26/130) | 11(11/104) | 0.07 | |
percentage (numbers of organs with metastasis / total organs inspected);
NA: data not available
P value was computed by Fisher exact test
4. Discussion
Currently, there are a limited number of known metastasis-suppressor genes in human CaP. Examples include KAI1, CD44, and MKK4 acting as metastasis-suppressor genes for CaP (Steeg, 2003;Vander Griend and Rinker-Schaeffer, 2004). Loss of these genes has been demonstrated during the clinical progression of the disease. MKK4 expression in human prostate cancers decreased with increasing Gleason grade (Steeg, 2003). Using a novel bioinformatic approach, we found decreased expression of SPARCL1 to be associated with CaP metastasis and demonstrate its ability to inhibit migration and invasiveness in vitro and metastasis in vivo.
SPARCL1 expression is found in most normal human tissues. High levels of SPARCL1 mRNA were found in prostate basal and columnar epithelium and cultured normal prostate epithelial cells. No expression was detected in a CaP cell line (LNCaP) or in immortalized poorly tumorigenic prostate epithelium (Nelson et al., 1998). The immunostained (IHC) for SPARCL1 staining pattern is also obtained in The Human Protein Atlas project (http://www.proteinatlas.org), in which six high-grade and four low-grade CaP samples are SPARCL1-negative while only one low grade CaP sample and a normal prostate sample are SPARCL1-positive. Furthermore, based on IHC analysis of human SPARCL1 expression on CaP tissue microarrays (TMAs), a recent study demonstrated a statistically significant inverse correlation between Gleason grades and SPARCL1 expression levels in 38 CaP patient samples (Hurley et al., 2012). There are multiple possible mechanisms for loss of SPARCL1 expression in cancer cells. Based on q-PCR of SPARCL1 gene copy numbers in PC3 cells, we found no SPARCL1 gene deletion in PC3 cells, but through data mining in public data sources (GEO and TCGA) we found that SPARCL1 locus is hyper-methylated in PC3 cells (GEO GSE12334) (Gal-Yam et al., 2008), breast cancer samples (TCGA) (Cancer Genome Atlas., 2012), and ovarian cancer samples (TCGA) (Cancer Genome Atlas., 2011).
SPARCL1 is a secreted protein that binds to collagen 1 to regulate collagen fibrillogenesis and results from a gene duplication of SPARC (Hambrock et al., 2003;Sullivan et al., 2006). SPARCL1 is closely related to the anti-adhesive extracellular matrix (ECM) protein known as BM-40 and SPARC. SPARC and SPARCL1 share 62% identity over C-terminal region with 232 amino acid residues but there is a greater difference in the highly acidic NH2-terminal domain of SPARCL1 (Girard and Springer, 1995). Although SPARC and SPARCL1 are related structurally and functionally, it is likely that the two proteins have both distinct and overlapping physiological roles (Lloyd-Burton and Roskams, 2012).
Using traditional in vitro approaches to study SPARCL1 function, a previous study on CaP cell lines demonstrated that SPARCL1 decreased the migratory and invasive properties of CaP cells but did not restrict the growth of prostate and CaP cells (Hurley et al., 2012). The data suggest that SPARCL1 may act as a suppressor of metastatic progression in prostate cancer. However, in vitro models do not accurately reflect in vivo metastasis and are unable to determine SPARCL1 function in vivo. Using an orthotopic mouse model for colon cancer, a recent study reported that SPARCL1 inhibited tumor growth in vivo and in vitro (Hu et al., 2012), suggesting that SPARCL1 functions as a tumor suppressor in colon cancer. Therefore, it remains unclear whether there is a suppressive role for SPARCL1 in CaP metastasis.
Our study is the first report to show the impact of SPARCL1 on CaP metastases in vivo and its impact on the overall survival of the mice. The two in vivo models (OX and IC injection) have been successfully used for studying metastasis genes in prostate cancer (Hafeez et al., 2012;Kim et al., 2003;Yang et al., 1999;Josson et al., 2011;Xu et al., 2006). The visceral metastasis patterns observed in the OX and IC models are similar (Table 1), indicating metastatic spread via common lymphatic or arterial circulation. SPARCL1 decreased metastatic progression significantly in both OX and IC models, suggesting an important contribution to late stages of extravasation and/or colonization.
Development of metastases was monitored by in vivo bioluminescence imaging (Bondareva et al., 2009). We found that whole-body (Fig. 4C and Fig. 5, lower right panel) photon emission rates (reflective of total tumor burden) were significantly diminished, suggesting that SPARCL1 can diminish distant PC3 invasiveness and reduce metastatic lesions in both visceral and skeletal sites. Because the frequencies of metastases at various anatomical sites were measured at the endpoint of each mouse (Table 1), our data represent final established metastatic lesions in late stage cancer rather than early metastatic development. Our results suggest that SPARCL1 was able to reduce the frequency of total visceral metastases (P ≤ 0.01) but had a smaller effect on the incidence of the development of bone metastases (P = 0.07).
Importantly, unlike SPARCL1’s role in colon cancer cells, SPARCL1 did not inhibit CaP cell proliferation in vitro (Fig. 3B, 3C, and Supplemental S1) but did decrease luciferase radiance in vivo (Fig. 4, Fig. 5 and Table 1). The radiance of luciferin signals directly derives from the expression level of active luciferase protein in PC3 cells. In our study, these signals correlated with primary tumor growth, local invasion, and metastases, but it was difficult to distinguish the signals of primary tumor growth from local invasion or metastases in vivo (Fig. 4A and Fig. 5). Therefore, we dissected primary prostate tumors and metastatic foci. We measured primary prostate tumor growth from moribund animals using luciferase radiances and tumor volumes of primary prostate tumors at sacrifice time (Supplemental Fig. S3). Our data reveal that there is a consistent pattern showing reduced prostate tumor volumes and luciferase radiance in SPARCL1 group in comparison with EV group at sacrifice time. However, the differences are not statistical significance (P > 0.05).
We realize that a major limitation in our study was the varying time of animal sacrifice which was dependent on moribundity in individual cases. Thus, time matched primary tumors from the EV and SPARCL1 groups were not available for tumor growth comparison. Therefore, while our data suggest that SPARCL1 does not decrease primary tumor growth, our data cannot exclude the possibility that SPARCL1 suppressed primary tumor growth in vivo, and SPARCL1 may play dual suppressive roles in primary tumor growth and metastasis.
The mechanism on SPARCL1 function is still largely unknown. In lung and pancreatic tumor xenografts, SPARCL1 is associated with desmoplasia (Brekken et al., 2004). In SPARCL1-null mice, the dermal elastic modulus was enhanced. The poor adhesive behavior of the fibroblasts cells on gels formed in the presence of SPARCL1 was due to alterations in fibril morphologies caused by SPARCL1, as opposed to a direct interaction of the cells with SPARCL1 (Sullivan et al., 2006). SPARC binds to collagen 1 to regulate collagen fibrillogenesis and assembly (Hambrock et al., 2003;Sullivan et al., 2006). A recent report suggests that SPARCL1 blocks the activation of the Ras homolog gene family, member C (RHOC), thereby inhibiting cellular movement (Hurley et al., 2012). It is possible that SPARCL1 might inhibit the activation of cancer associated fibroblasts (CAF) (Kalluri and Zeisberg, 2006) through remodeling of cancer extracellular matrix (ECM) (Ramos et al., 1998). The aforementioned mechanisms might be responsible for SPARCL1 suppressive function in CaP, explaining how overexpression of SPARCL1 in PC-3 cells compared to control cells confers a two-week survival advantage for the mice (Fig. 4).
In conclusion, we have provided evidence that SPARCL1 suppresses metastasis in CaP. This study sets the stage for further investigations on the basic mechanisms that underlie cancer metastasis. Additional studies on SPARCL1 will be valuable for determining its mechanisms of metastasis suppression in cancer.
Supplementary Material
Cells were cultured either with SPARCL1 (10 μg/ml, closed circles) or without SPARCL1 (control, open circles). After certain days incubation as indicated in X-axis, SPARCL1 effect of cellular proliferation was assessed. The proliferation rates at different time points are presented as means S.E. (n=4) in Y-axis. The proliferation differences over three days for PC3 and ARCaPM were non-significant between SPARCL1 treated cells and control cells (p > 0.05; ANOVA).
To monitor orthotopic prostate tumor spread to other organs, the end-point metastatic tumors were dissected and imaged. Images of organ luciferase are listed from orthotopic xenograft mice bearing PC3-luc/EV (left) and PC3-luc/SPARCL1 (right) tumors. All images were set using the same pseudocolor scale in radiance.
(A) Mean tumor bioluminescence index was compared as a bar graph between the control PC3-luc/EV mice and the PC3-luc/SPARCL1 mice (n = 8). Primary prostate luminescence index (Log 10) is displayed on the Y-axis to represent average tumor growth at sacrifice.
(B) Measurements of mean in vivo orthotopic tumor volume at sacrifice time point. Mean tumor volumes from mice bearing PC3-luc/EV and PC3-luc/SPARCL1 tumors (n = 8) were determined and plotted in a bar graph. The bar heights at the Y-axis represent final orthotopic tumor volume (cm3).
Note: Either the mean tumor bioluminescence index (panel A) or the tumor volume (panel B) between PC3-luc/EV and PC3-luc/SPARCL1 groups was compared on the X-axis using a Student’s t test. *NS: nonsignificant
Research highlights.
We identify a candidate suppressor gene (SPARCL1) from a cancer prognostic signature.
We examine expression patterns of SPARCL1 in human prostate tissues and cell lines.
Overexpression of SPARCL1 does not affect tumor cell growth in vitro.
SPARCL1 suppresses tumor cell migration, invasiveness, and metastasis.
Acknowledgments
Grant Support: This work was supported in part by a Howard Temin Award from the National Cancer Institute at the National Institutes of Health (CA114033 to YY), American Cancer Society-Institutional Research Grant (#IRG-58-009-51 and #IRG-58-009-53), and the Vanderbilt Clinical and Translational Science Awards (CTSA) UL1 RR024975 from National Center for Research Resources (NCRR), a part of the National Institutes of Health (NIH), (CRC1838 to YY), and the National Cancer Institute (4R01-CA076142-14 to RJM). DJD was supported by the American Cancer Society Great Lakes Division-Michigan Cancer Research Fund Postdoctoral Fellowship.
The authors thank Dr. Harold L Moses and Dr. Jennifer Pietenpol for assistance in experiments; Dr. K. Pienta for providing bioluminescent human prostate carcinoma cell line (PC3-Luc); The Vanderbilt Cooperative Human Tissue Network for human tissue samples.
Abbreviations
- CaP
cancer of the prostate gland
- H.E
hematoxylin and eosin
- IC
Intracardiac
- IHC
Immunohistochemistry
- IVIS
in vivo Imaging System
- OX
orthotopic xenografting
- PC3-Luc
the bioluminescent human prostate carcinoma cell line
- PC3-luc/EV
GFP-positive PC3-Luc cells expressing empty control vector
- PC3-luc/SPARCL1
GFP-positive PC3-Luc cells overexpressing SPARCL1
- SCID
Severe combined immunodeficient
- SPARCL1
secreted protein acidic and rich in cysteine-like 1
Footnotes
Disclosure/Conflict of interest
The authors declare no disclosure or conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yuzhu Xiang, Email: yuzhuxiang@gmail.com.
Qingchao Qiu, Email: qingchao.qiu@vanderbilt.edu.
Ming Jiang, Email: ming.jiang.1@ntu.edu.cn.
Renjie Jin, Email: renjie.jin@Vanderbilt.edu.
Brian D Lehmann, Email: brian.d.lehmann@Vanderbilt.edu.
Douglas W Strand, Email: doug.strand@Vanderbilt.edu.
Bojana Jovanovic, Email: bojana.jovanovic@Vanderbilt.edu.
David J DeGraff, Email: david.degraff@Vanderbilt.edu.
Yi Zheng, Email: milozhengyi@gmail.com.
Dina A Yousif, Email: dina.a.yousif@Vanderbilt.edu.
Thomas C Case, Email: tom.case@Vanderbilt.edu.
Jia Yi, Email: jyi5@uthsc.edu.
Justin M Cates, Email: justin.m.cates@Vanderbilt.edu.
John Virostko, Email: jack.virostko@vanderbilt.edu.
Xiusheng He, Email: hexiusheng@hotmail.com.
Xunbo Jin, Email: jinxunbo@163.com.
Simon W Hayward, Email: simon.hayward@vanderbilt.edu.
Robert J Matusik, Email: robert.matusik@vanderbilt.edu.
Alfred L George, Jr, Email: al.george@vanderbilt.edu.
References
- Ashby WJ, Wikswo JP, Zijlstra A. Magnetically attachable stencils and the nondestructive analysis of the contribution made by the underlying matrix to cell migration. Biomaterials. 2012;33:8189–8203. doi: 10.1016/j.biomaterials.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendik I, Schraml P, Ludwig CU. Characterization of MAST9/Hevin, a SPARC-like protein, that is down-regulated in non-small cell lung cancer. Cancer Res. 1998;58:626–629. [PubMed] [Google Scholar]
- Bondareva A, Downey CM, Ayres F, Liu W, Boyd SK, Hallgrimsson B, Jirik FR. The lysyl oxidase inhibitor, beta-aminopropionitrile, diminishes the metastatic colonization potential of circulating breast cancer cells. PLoS One. 2009;4:e5620. doi: 10.1371/journal.pone.0005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekken RA, Sullivan MM, Workman G, Bradshaw AD, Carbon J, Siadak A, Murri C, Framson PE, Sage EH. Expression and characterization of murine hevin (SC1), a member of the SPARC family of matricellular proteins. J Histochem Cytochem. 2004;52:735–748. doi: 10.1369/jhc.3A6245.2004. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cher ML, de Oliveira JG, Beaman AA, Nemeth JA, Hussain M, Wood DP., Jr Cellular proliferation and prevalence of micrometastatic cells in the bone marrow of patients with clinically localized prostate cancer. Clin Cancer Res. 1999;5:2421–2425. [PubMed] [Google Scholar]
- Clark CJ, Sage EH. A prototypic matricellular protein in the tumor microenvironment--where there’s SPARC, there’s fire. J Cell Biochem. 2008;104:721–732. doi: 10.1002/jcb.21688. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Esposito I, Kayed H, Keleg S, Giese T, Sage EH, Schirmacher P, Friess H, Kleeff J. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia. 2007;9:8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard JP, Springer TA. Cloning from purified high endothelial venule cells of hevin, a close relative of the antiadhesive extracellular matrix protein SPARC. Immunity. 1995;2:113–123. doi: 10.1016/1074-7613(95)90083-7. [DOI] [PubMed] [Google Scholar]
- Girard JP, Springer TA. Modulation of endothelial cell adhesion by hevin, an acidic protein associated with high endothelial venules. J Biol Chem. 1996;271:4511–4517. doi: 10.1074/jbc.271.8.4511. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Hafeez BB, Zhong W, Fischer JW, Mustafa A, Shi X, Meske L, Hong H, Cai W, Havighurst T, Kim K, Verma AK. Plumbagin, a medicinal plant (Plumbago zeylanica)-derived 1,4-naphthoquinone, inhibits growth and metastasis of human prostate cancer PC-3M-luciferase cells in an orthotopic xenograft mouse model. Mol Oncol. 2012 doi: 10.1016/j.molonc.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambrock HO, Nitsche DP, Hansen U, Bruckner P, Paulsson M, Maurer P, Hartmann U. SC1/hevin. An extracellular calcium-modulated protein that binds collagen. I J Biol Chem. 2003;278:11351–11358. doi: 10.1074/jbc.M212291200. [DOI] [PubMed] [Google Scholar]
- Hendriksen PJ, Dits NF, Kokame K, Veldhoven A, van Weerden WM, Bangma CH, Trapman J, Jenster G. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Res. 2006;66:5012–5020. doi: 10.1158/0008-5472.CAN-05-3082. [DOI] [PubMed] [Google Scholar]
- Hu H, Zhang H, Ge W, Liu X, Loera S, Chu P, Chen H, Peng J, Zhou L, Yu S, Yuan Y, Zhang S, Lai LL, Yen YCD, Zheng S. Secreted Protein Acidic and Rich in Cysteines-Like 1 Suppresses Aggressiveness and Predicts Better Survival in Colorectal Cancers. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0124. [DOI] [PubMed] [Google Scholar]
- Hurley PJ, Marchionni L, Simons BW, Ross AE, Peskoe SB, Miller RM, Erho N, Vergara IA, Ghadessi M, Huang Z, Gurel B, Park BH, Davicioni E, Jenkins RB, Platz EA, Berman DM, Schaeffer EM. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jiang M, Strand DW, Fernandez S, He Y, Yi Y, Birbach A, Qiu Q, Schmid J, Tang DG, Hayward SW. Functional remodeling of benign human prostatic tissues in vivo by spontaneously immortalized progenitor and intermediate cells. Stem Cells. 2010;28:344–356. doi: 10.1002/stem.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josson S, Nomura T, Lin JT, Huang WC, Wu D, Zhau HE, Zayzafoon M, Weizmann MN, Gururajan M, Chung LW. beta2-microglobulin induces epithelial to mesenchymal transition and confers cancer lethality and bone metastasis in human cancer cells. Cancer Res. 2011;71:2600–2610. doi: 10.1158/0008-5472.CAN-10-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 2003;169:1122–1133. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Johnson M, Koterba K, Herynk MH, Uehara H, Gallick GE. Reduced c-Met expression by an adenovirus expressing a c-Met ribozyme inhibits tumorigenic growth and lymph node metastases of PC3-LN4 prostate tumor cells in an orthotopic nude mouse model. Clin Cancer Res. 2003;9:5161–5170. [PubMed] [Google Scholar]
- Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, Albertini V, Malek A, Chiorino G, Catapano CV, Carbone GM. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Burton S, Roskams AJ. SPARC-like 1 (SC1) is a diversely expressed and developmentally regulated matricellular protein that does not compensate for the absence of SPARC in the CNS. J Comp Neurol. 2012;520:2575–2590. doi: 10.1002/cne.23029. [DOI] [PubMed] [Google Scholar]
- Nelson PS, Plymate SR, Wang K, True LD, Ware JL, Gan L, Liu AY, Hood L. Hevin, an antiadhesive extracellular matrix protein, is down-regulated in metastatic prostate adenocarcinoma. Cancer Res. 1998;58:232–236. [PubMed] [Google Scholar]
- Park SI, Kim SJ, McCauley LK, Gallick GE. Pre-clinical mouse models of human prostate cancer and their utility in drug discovery. Curr Protoc Pharmacol. 2010;Chapter 14(Unit) doi: 10.1002/0471141755.ph1415s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P, McMillan TJ. Use of the tetrazolium assay in measuring the response of human tumor cells to ionizing radiation. Cancer Res. 1990;50:1392–1396. [PubMed] [Google Scholar]
- Qiu QC, Lu PC, Xiang YZ, Shyr Y, Chen X, Lehmann BD, Viox DJ, George AL, Jr, Yi Y. A data similarity-based strategy for meta-analysis of transcriptional profiles in cancer. PLoS One. 2013;8:e54979. doi: 10.1371/journal.pone.0054979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos DM, Chen B, Regezi J, Zardi L, Pytela R. Tenascin-C matrix assembly in oral squamous cell carcinoma. Int J Cancer. 1998;75:680–687. doi: 10.1002/(sici)1097-0215(19980302)75:5<680::aid-ijc4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- Sullivan MM, Barker TH, Funk SE, Karchin A, Seo NS, Hook M, Sanders J, Starcher B, Wight TN, Puolakkainen P, Sage EH. Matricellular hevin regulates decorin production and collagen assembly. J Biol Chem. 2006;281:27621–27632. doi: 10.1074/jbc.M510507200. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Griend DJ, Rinker-Schaeffer CW. A new look at an old problem: the survival and organ-specific growth of metastases. Sci STKE. 2004;2004:e3. doi: 10.1126/stke.2162004pe3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xia XQ, Jia Z, Sawyers A, Yao H, Wang-Rodriquez J, Mercola D, McClelland M. In silico estimates of tissue components in surgical samples based on expression profiling data. Cancer Res. 2010;70:6448–6455. doi: 10.1158/0008-5472.CAN-10-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Peterse JL, van’ V. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- Wu J, Qiu Q, Xie L, Fullerton J, Yu J, Shyr Y, George AL, Jr, Yi Y. Web-based interrogation of gene expression signatures using EXALT. BMC Bioinformatics. 2009;10:420. doi: 10.1186/1471-2105-10-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang R, Xie ZH, Odero-Marah V, Pathak S, Multani A, Chung LW, Zhau HE. Prostate cancer metastasis: role of the host microenvironment in promoting epithelial to mesenchymal transition and increased bone and adrenal gland metastasis. Prostate. 2006;66:1664–1673. doi: 10.1002/pros.20488. [DOI] [PubMed] [Google Scholar]
- Yang M, Jiang P, Sun FX, Hasegawa S, Baranov E, Chishima T, Shimada H, Moossa AR, Hoffman RM. A fluorescent orthotopic bone metastasis model of human prostate cancer. Cancer Res. 1999;59:781–786. [PubMed] [Google Scholar]
- Yang SW, Chanda D, Cody JJ, Rivera AA, Waehler R, Siegal GP, Douglas JT, Ponnazhagan S. Conditionally replicating adenovirus expressing TIMP2 increases survival in a mouse model of disseminated ovarian cancer. PLoS One. 2011;6:e25131. doi: 10.1371/journal.pone.0025131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Li C, Miller C, George AL., Jr Strategy for encoding and comparison of gene expression signatures. Genome Biol. 2007;8:R133. doi: 10.1186/gb-2007-8-7-r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SJ, Yu JK, Ge WT, Hu HG, Yuan Y, Zheng S. SPARCL1, Shp2, MSH2, E-cadherin, p53, ADCY-2 and MAPK are prognosis-related in colorectal cancer. World J Gastroenterol. 2011;17:2028–2036. doi: 10.3748/wjg.v17.i15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Zaravinos A, Lambrou GI, Boulalas I, Delakas D, Spandidos DA. Identification of common differentially expressed genes in urinary bladder cancer. PLoS One. 2011;6:e18135. doi: 10.1371/journal.pone.0018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells were cultured either with SPARCL1 (10 μg/ml, closed circles) or without SPARCL1 (control, open circles). After certain days incubation as indicated in X-axis, SPARCL1 effect of cellular proliferation was assessed. The proliferation rates at different time points are presented as means S.E. (n=4) in Y-axis. The proliferation differences over three days for PC3 and ARCaPM were non-significant between SPARCL1 treated cells and control cells (p > 0.05; ANOVA).
To monitor orthotopic prostate tumor spread to other organs, the end-point metastatic tumors were dissected and imaged. Images of organ luciferase are listed from orthotopic xenograft mice bearing PC3-luc/EV (left) and PC3-luc/SPARCL1 (right) tumors. All images were set using the same pseudocolor scale in radiance.
(A) Mean tumor bioluminescence index was compared as a bar graph between the control PC3-luc/EV mice and the PC3-luc/SPARCL1 mice (n = 8). Primary prostate luminescence index (Log 10) is displayed on the Y-axis to represent average tumor growth at sacrifice.
(B) Measurements of mean in vivo orthotopic tumor volume at sacrifice time point. Mean tumor volumes from mice bearing PC3-luc/EV and PC3-luc/SPARCL1 tumors (n = 8) were determined and plotted in a bar graph. The bar heights at the Y-axis represent final orthotopic tumor volume (cm3).
Note: Either the mean tumor bioluminescence index (panel A) or the tumor volume (panel B) between PC3-luc/EV and PC3-luc/SPARCL1 groups was compared on the X-axis using a Student’s t test. *NS: nonsignificant