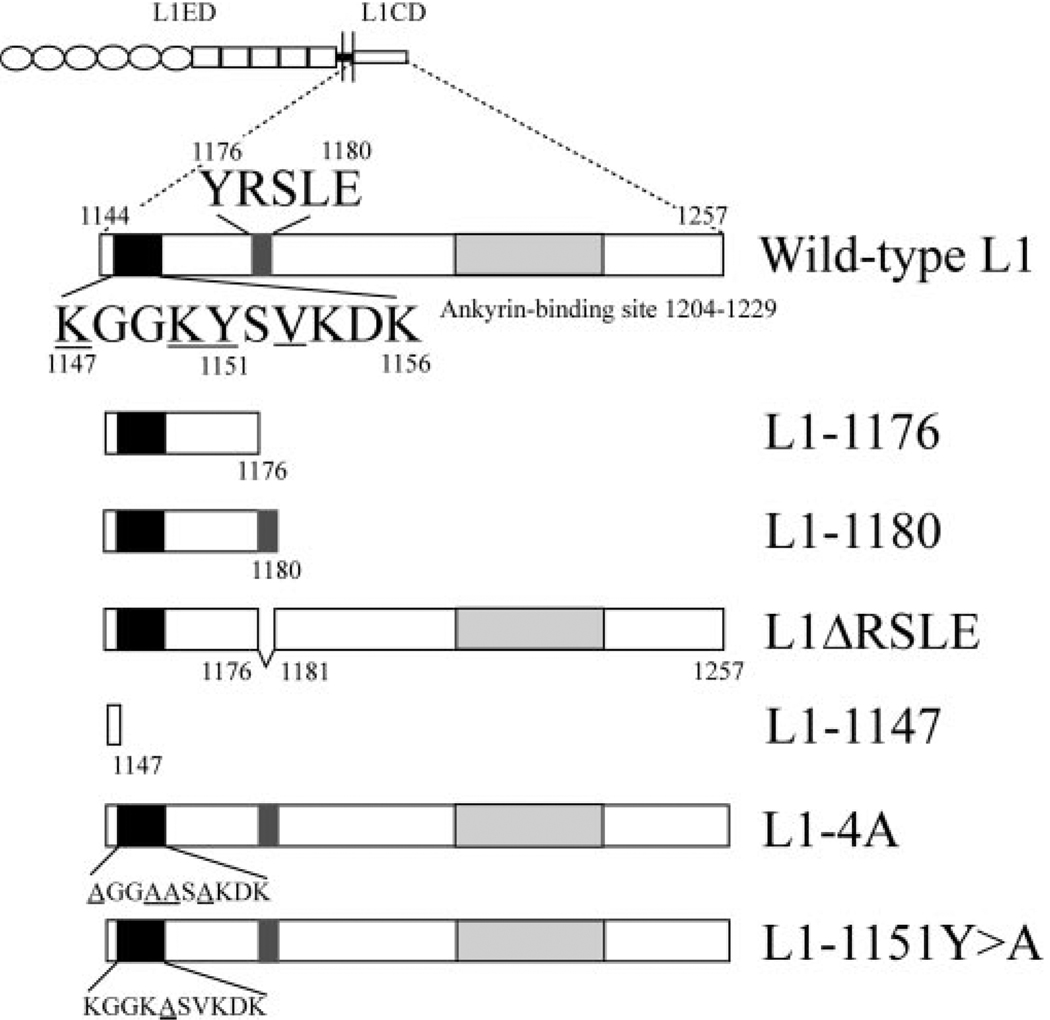
Figure 2.
Schematic demonstration of the L1 intracellular mutations. Number of the amino acids in the L1CD of wild-type L1 (aa 1144–1257) is numbered by their position in the open reading frame of the human L1 gene. The juxtamembrane region, the YRSLE sequence, and the ankyrin-binding sites are highlighted. The numbers of the key residues are indicated on top or at the bottom of the corresponding residue. The underlined residues at the juxtamembrane region are predicted to be the ERM binding site by homology alignment to ICAM-2. The L1-1176 construct is truncated after the Y1176 residue. The L1-1147 construct is truncated after the K1147 residue. The L1-1180 residue is truncated after the E1180 residue. The L1ΔRSLE construct has an internal deletion from the R1177 to E1180 residue. The juxtamembrane mutants L1-4A and L1-1151Y>A change critical residues in the juxtamembrane region to alanine. The mutated residues are underlined.