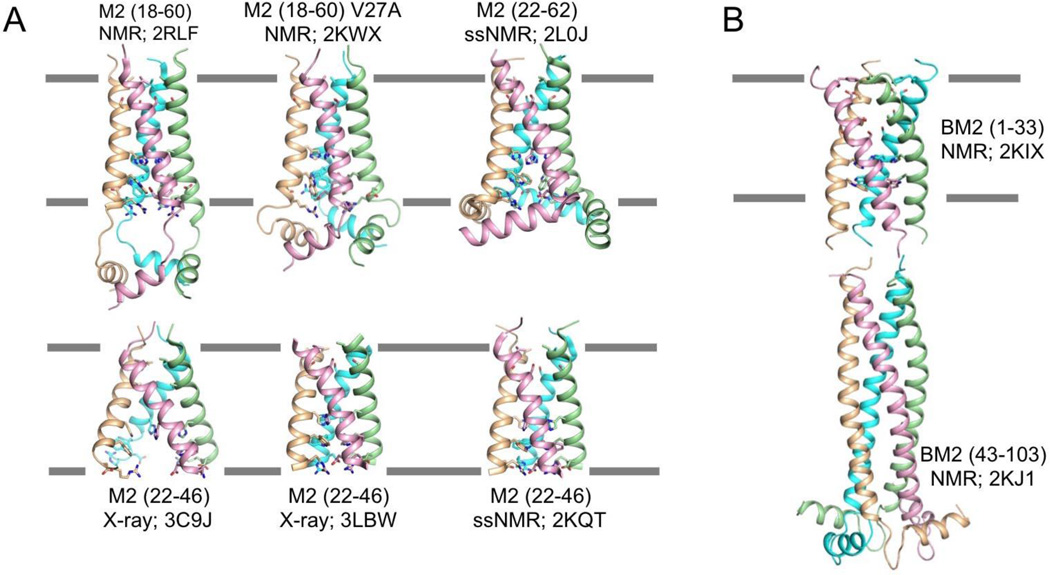
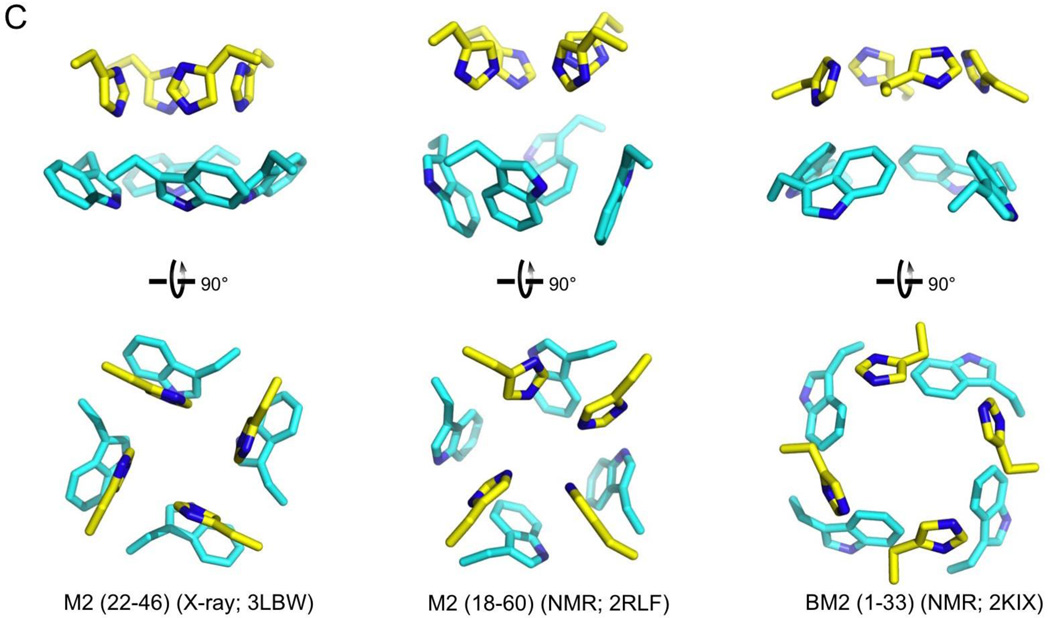
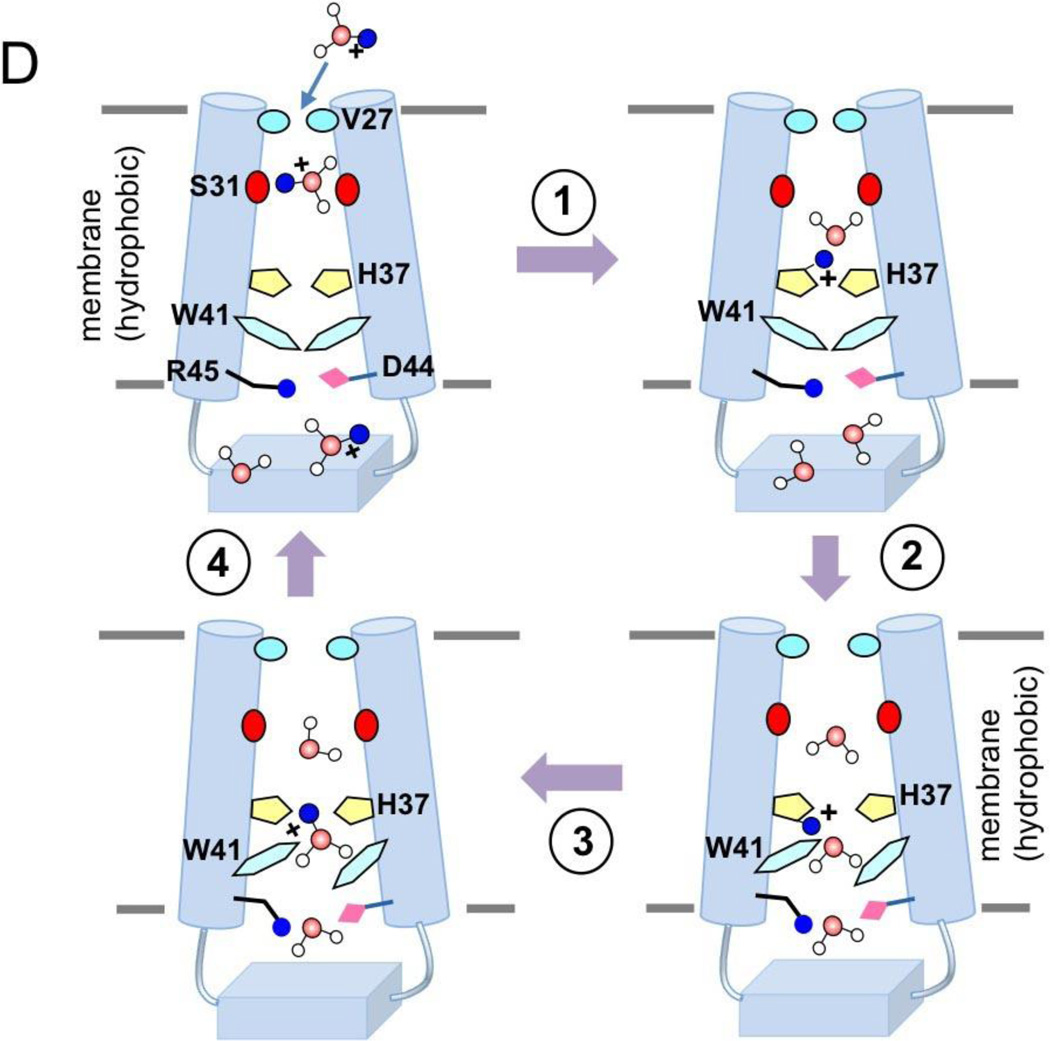
Figure 1. Structures of the influenza proton channels and mechanism of proton conduction.
(A) The many structures of the influenza M2 channel. The PDB codes 2RLF and 2KWX represent the solution NMR structures of the wildtype and the V27A mutant determined using residues 18–60. The 3C9J and 3LBW are crystal structures of the TM domain (residues 22–46) determined at pH 7.3 and pH 6.5, respectively. The structures 2L0J and 2KQT were obtained using solid-state NMR using protein constructs that encompass residues 22–62 and residues 22-46, respectively. (B) Solution NMR structures of the BM2 protein. The PDB codes 2KIX and 2KJ1 represent the structures of the TM domain (residues 1-33) and the cytoplasmic domain (residues 43-103), respectively.
(C) Isolated view of the pore-lining histidine and tryptophan sidechains in M2 and BM2 channels. Images are from the high resolution crystal structure (3LBW) and NMR structure (2RLF) of M2 and NMR structure of BM2 (2KIX).
(D) Cartoon representation of the mechanism of proton conduction. (1) Channel breathing due to thermal energy can transiently enlarge the N-terminal opening, allowing protons to be relayed cross the Val27 barrier. (2) The protons bind to His37 imidazole, and protonation of His37 triggers opening of the Trp41 gate. (3) C-terminal water molecules accept protons from protonated His37. (4) Polar residues Asp44 and Arg45 facilitate proton exit.