Abstract
Scleroderma (SSc) is a complex and heterogeneous connective tissue disease mainly characterized by autoimmunity, vascular damage and fibrosis that mostly involve the skin and lungs. Epstein-Barr virus (EBV) is a lymphotropic-γ-Herpesvirus that has co-evolved with human species infecting >95% of the adult population worldwide, and has been a leading candidate in triggering several autoimmune diseases. Here we show that EBV establishes infection in the majority of fibroblasts and endothelial cells in the skin of SSc patients, characterized by the expression of the EBV-non-coding-small-RNAs (EBER) and the increased expression of immediate-early-lytic and latency mRNAs and proteins. We report that EBV is able to persistently infect human-SSc-fibroblasts in vitro, inducing an aberrant innate immune response in infected cells. EBV-TLR aberrant activation induces the expression of selected IRFs, ISGs, TGFβ1 and several markers of fibroblast activation, such as smooth muscle actin and Endothelin-1, and all of these genes play a key role in determining the pro-fibrotic phenotype in SSc-fibroblasts.
These findings imply that EBV infection occurring in mesenchymal, endothelial, and immune cells of SSc patients may underlie the main pathological features of SSc including autoimmunity, vasculopathy and fibrosis, and provide a unified disease mechanism represented by EBV reactivation.
Introduction
Fibrosis is a pathologic scarring process that has been considered the hallmark of systemic sclerosis (scleroderma) (SSc), a connective tissue disease characterized by autoimmunity, inflammation and vasculopathy, leading to progressive fibrosis mostly of skin and lungs. Fibroblasts, mainly myofibroblasts, clearly have a necessary and fundamental role in promoting fibrosis (Varga and Abraham, 2007). Evidence suggests that innate immune activation of Interferon (IFNs) by Tolllike receptors (TLRs) may play a role in the pathogenesis of inflammation in many autoimmune diseases, initially assessed in systemic lupus erythematous (SLE) and more recently in SSc, implying that dysregulation of the innate immune response underlies the overactive immune system of individuals who are susceptible to autoimmune disease (Theofilopoulos, 2012).
Recently it has been shown by our group and by other independent studies that IFN signature gene expression is increased in peripheral blood mononuclear cells (PBMCs) and in the skin of SSc patients (Farina et al., 2010a; York et al., 2007). Further we showed that dsRNA/Poly(I:C), a TLR3 ligand, stimulates interferon production, inflammation and markers of vascular activation in SSc skin (Farina et al., 2011; Farina et al., 2010b).
While our studies strongly supported the potential key role for dsRNA/Poly(I:C) to contribute to innate immune activation in SSc, the origin of the innate immune dysregulation in SSc is unknown, and to date there is no obvious evidence addressing any potential exogenous/endogenous source of RNA that might represent a TLR3-ligand in SSc skin. In order to address this question, we asked if viral RNAs could be detected in the skin and serve as innate immune ligands in SSc. Previous studies in animal models have shown that murine Cytomegalovirus-infection in immuno-compromised mice resembles the pathological processes seen in autoimmune diseases, particularly in SSc, suggesting a link between Herpesvirus-infection and the development of fibrosis (Pandey and LeRoy, 1998). However no direct evidence of CMV-infection, such as the presence of viral protein or production of viral progeny, has yet been found in SSc. Since Herpesvirus-family has been linked to development of fibrosis we focused our attention on Epstein-Barr virus (EBV), a γ-Herpesvirus that has been a leading candidate in triggering several autoimmune diseases (Dreyfus, 2011; Ebrahimi et al., 2001; Niller et al., 2008; Stoolman et al., 2011). This virus is a biologically plausible source for endogenous innate immune activation since it is ubiquitous in nature, establishes a lifelong silent infection with continuous virus production due to reactivation, and most importantly, modulates the human immune system, evolving immune evasion strategies to the host antiviral response (Martorelli et al., 2012; Munz et al., 2009; Young and Rickinson, 2004).
We show that EBV is able to infect human-dermal-fibroblasts, and modulate the innate immune response in infected-fibroblasts, inducing fibroblast-myofibroblast conversion typical of a pro-fibrotic phenotype. We further demonstrate the presence of EBV viral transcripts and proteins in the majority of fibroblasts and endothelial cells in the skin of SSc patients supporting a crucial role of EBV in SSc pathogenesis. Viral infection of non-immune cells might provide a persistent source of tissue injury and induce chronic inflammation and fibrosis in SSc skin.
Results
EBV transcripts are present in skin and in peripheral blood mononuclear cells (PBMCs) from SSc patients
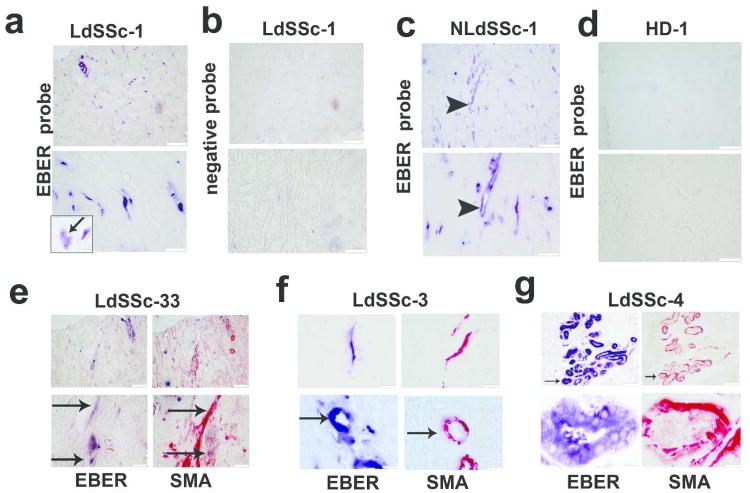
The EBV non coding RNA, Epstein-Barr virus small RNA (EBER) is the most abundant viral transcript in latently infected cells, and can activate TLR3 and RIG-I (Iwakiri et al., 2009; Samanta et al., 2008). We investigated EBER (EBER1/EBER2) expression in SSc skin by “in situ” hybridization (ISH) in sections from representative SSc patients (demographics and clinical characteristics are specified in Table S1 and S2). We found a striking accumulation of EBER-positive-cells (EBER+cells) in the deep dermis of lesional/forearm (LdSSc) and non-lesional/back (NLdSSc) SSc skin (Fig. 1a,c). EBER+cells were distributed among the bundles of the extracellular matrix (ECM) in the deep dermis, showing spindle morphology and identified as fibroblasts by the shape and location. The EBER signal was mostly detected in cell nuclei, although certain nuclei of those cells appear mostly destroyed and disintegrated (Fig. 1a). No EBER+cells were detected in healthy donor (HD) skin (Fig. 1d, Table S2). Since myofibroblasts play a pivotal role in fibrotic SSc skin, we asked whether EBER+cells may express smooth-muscle-actin (SMA), a marker for myofibroblasts. Immunohistochemical staining (IHC) of serial skin biopsies showed that most, but not all SMA-stained-cells co-localizied with EBER+cells (Fig. 1e,f), Interestingly EBER+cells were also detected around blood vessels, identified as smooth-muscle-cells (SMCs) by SMA IHC (Fig. 1f, lower-panels). We also observed EBER+cells in the myo-epithelial layer of the cutaneous glands (Fig. 1g), as well as in most of the endothelial cells in dermal ectatic vessels (Fig. 1c). No EBER staining was detected in SMCs, myo-epithelial or endothelial cells from HDs (Fig. S1). EBER+fibroblasts were also found in patients with limited SSc (lSSc) (data not shown).
Figure 1. EBER infected cells in SSc skin.
(a-c) Representative images of in situ hybridization (ISH) in lesional (LdSSc-1) and in non/lesional diffuse skin (NLdSSc-1), (a) showing EBER+spindle-cells in the deep-dermis; (b) EBER-staining in LdSSc-1 skin with probe-control; (c) EBER-staining in endothelial cells of ectasic vessels in the skin; (d) EBER-staining of healthy donor (HD) skin; (upper-panels scale/bar 100μm, lower-panels 50μm). (e-f, upper-panel) Co-localization of EBER+cells detected by ISH (blue) with α-smooth muscle actin (αSMA) cells (red) detected by IHC in serial LdSSc skin sections (bar/scale 100μm and 50μm); (f lower-panel) EBER+staining in blood vessels smooth muscle cells identified by αSMA staining by IHC in serial skin section (lower-panel, bar/scale 50μm). (g) EBER+cells detected in myo-epithelial-glands surrounded by SMA fibers in skin (bar/scale 50μm).
It is known that in B-cells detection of EBV DNA and EBER or lytic RNAs/proteins identifies latent or active infection, respectively (Okano et al., 2005). Thus, we asked if EBV infection is associated with expression of lytic- or latency-genes in SSc skin. We found mRNA expression of both BZLF1, the viral-transactivator that drives EBV reactivation, switching from latency to the lytic-replication, and EBNA1, one of the 9 latency-genes, present in LdSSc skin (Fig. S2a). cDNA sequencing confirmed BZLF1 and EBNA1 specificity of the RT-PCR products (data not shown). BZLF1 was also detected in NLdSSc and in none of the control skin (Fig. S2b) (Table S2).
As EBV persists in B-cells, we also investigated the state of EBV infection in SSc PBMCs. We found that BZLF1 gene, lytic- and latency-proteins were significantly increased in PBMCs from SSc patients (Fig. S2c, Table S2,S7). While all the patients and HDs were seropositive for EBV, a striking increased production of antibodies against EBV-viral-capsid-antigen (VCA), the marker of acute EBV-infection was detected in SSc sera (Tables S1 and S3).
EBV DNA is detected in the skin of SSc patients
To further confirm the presence of EBV infection in SSc skin we measured EBV genome DNA. We found EBV-EBER1 in LdSSc (Fig. S2d) and NLdSSc skin but rarely in HD skin (Table S2). DNA sequencing confirmed that amplified DNA was indeed EBER1 (data not shown). Increased copies of EBV-DNA were found in LdSSc compared to HD skin (Fig. S2e). Since it has been shown that most of the immunosuppressive medications used to treat organ transplant recipients elevate EBV load and reactivate lytic/latency-genes as has also been shown in patients with SLE or other autoimmune disease, we selected a group of SSc patients naïve for any treatment compared to SSc patients who received immunosuppressive therapy (Green et al., 2000; Larsen et al., 2011). We found no statistical differences in EBV-DNA viral load (EBER1) and EBER expression in the skin of the treatment naïve SSc patients compared to the treated group (Fig. S2f and S4a-c) (Table S4-5). Undetectable levels of EBV-DNA were measured in the skin from foreskin samples as well as in 293-cells as negative control.
Assessment of viral proteins in SSc skin
We next investigated whether EBV transcripts might produce viral proteins in SSc skin. Comparable to data obtained with EBV transcripts, we found that the majority of the SSc patients positive for EBV mRNA showed expression of lytic- as well as latent-proteins in the skin (Fig. S3a-f, Table S6-7). Noteworthy, no lymphocyte infiltration was observed in areas where ZEBRA positive cells were detected. Nuclei of endothelial-cells and SMCs were also positive for BFLF2 and ZEBRA (Fig. S3c). No difference in ZEBRA protein skin expression was observed between biopsies from patient's naïve to immunosuppressed treatment and biopsies from patients treated with immunosuppression (Fig. S4d-f). Notably, we did not find expression of the late/EBV-protein gp350/220 (Fig. S5), or detection of mature virus production by electron microscopy in SSc skin (data not shown), suggesting that the virus lytic cycle is abortive (Martel-Renoir et al., 1995). None of the HD skin sections showed specific staining for ZEBRA and BFLF2 (Fig. S5).
EBV infects human SSc-fibroblasts in vitro
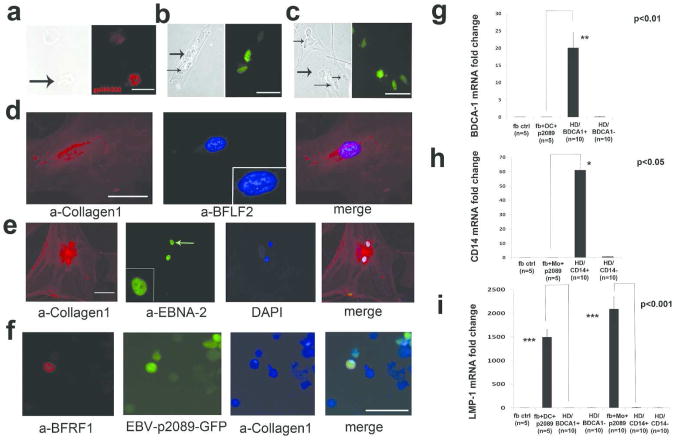
EBV mainly infects B lymphocytes through CD21 receptor, although it is generally accepted that it also infects epithelial cells, even though they are CD21-negative. Thus its presence in fibroblasts was unexpected. No expression of EBV DNA, mRNA and/or protein was detected in primary-fibroblast-cell-lines. The first attempt of using cell-free-EBV or recombinant-EBV-p2089 (EBV-p2089), failed to infect any fibroblast cell lines and it prompted us to explore how the virus gains access into fibroblasts. Since monocytes (MO) have been found to be increased in the perivascular areas of SSc skin, we used purified MO or dendritic cells (DCs) from HDs bound with EBV-p2089 to perform transfer-infection, as indicated by previous reports using resting B-cells as a transfer vehicle for EBV-infection of epithelial-cells (Feederle et al., 2007; Shannon-Lowe et al., 2006; York et al., 2007). MO or DCs-virus-binding efficiency was detected by IFA using EBV-anti-capsid-ab (10-20% of MO or DCs were able to bind the virus) (Fig. 2a). No EBV infection was detected in MO/DCs recovered after fibroblast-transfer-infection. After the transfer-infection 10% cell/field of SSc-fibroblasts showed nuclear p2089-GFP-fluorescence at 5 days and 4 weeks post-infection (PI), respectively (Fig. 2b-c and Fig. S6a). Since fibroblasts do not represent the natural target of EBV, we analyzed the expression of latency as well as lytic-proteins, in order to identify the viral strategy in this EBV-infected-cell-type. EBV lytic-proteins, BFLF2/BFRF1, and latent-antigen/EBNA2 were expressed at 4 weeks PI in the nuclei of infected-SSc-cells defined as fibroblasts by collagen-1 and collagen-11 staining and by shape (Fig. 2d-f and data not shown). We did not find the expression of late-lytic-gene/BLLF1, suggesting that abortive EBV-replication occurs in EBV-infected-SSc-fibroblasts (data not shown). We did not find any cleavage of PARP protein in EBV-p2089- compared to mock-infected-SSc-fibroblasts, confirming that EBV does not induce apoptosis in infected-SSc-cells, as well as in infected-B-cells (Fig. S6b).
Figure 2. In vitro infection of SSc fibroblast.
Monocytes (MO), Dendritic Cells (DCs) from healthy donors (HDs) previously exposed to EBV-p2089 were co-cultured with SSc-fibroblasts as described in the methods. (a) Indirect-immunofluorescence-staining (IF) for EBV-gp350/220-antigen of MO bound to EBV-p2089 (left-panel: phase-contrast-light-microcopy, bar/scale 20μm). (b and c) Detection of EBV-p2089-GFP in fibroblasts after 5 days (b) and 4/week (c) post-infection (left panel: phase-contrast-light-microcopy, bar/scale 20μm). (d -e) Double-IF of adherent or in suspension fibroblasts (f) co-stained with the indicated antibodies after 4/week post-infection (square-insert bar-scale 10μm). Diaminidino-2-phenylindole (DAPI) was used as counterstaining for the nuclei. (g-i) mRNA expression of indicated genes in p2089-fibroblasts-transfer-infected-cultures, CD14+/CD14- and BDCA1+/BDCA1-population from HDs by q-PCR. Data are expressed as the fold-change normalized to mRNA expression in a single sample from HD. Bars represent mean±S.E.M.
MO/DCs derived from different HDs consistently infected all SSc-fibroblast-cell-lines included in this study. Infected SSc-fibroblast-cultures were monitored by IFA. Fibroblasts from NLdSSc and from HDs were also occasionally infected (ratio of 1 of 4). EBV-p2089-DNA persisted up to six months in infected LdSSc-fibroblasts, while EBV-p2089-infected-NLdSSc-fibroblasts and EBV-p2089-infected-HD-fibroblasts died after 20 days PI.
To exclude immune cell contamination in the fibroblast population, markers of MO and DCs were evaluated by qPCR. CD14 and BDCA1 mRNA expression were undetectable in MO, or DCs transfer-infected- or mock-infected-fibroblasts, although those markers were expressed in MO and DCs used as shuttle-infection in fibroblast cultures (Fig. 2g-h). Interestingly, LMP1 mRNA expression was found significantly abundant in EBV-infected-SSc-cells, whereas no LMP1 expression was detected in uninfected-,mock-infected or MO/DCs from HDs, confirming that EBV is infecting SSc-fibroblasts (Fig. 2i). IFA staining for CD14/BDCA1 was absent in infected-SSc-fibroblasts (data not shown).
EBV activates TLR pathway in infected-SSc-fibroblasts
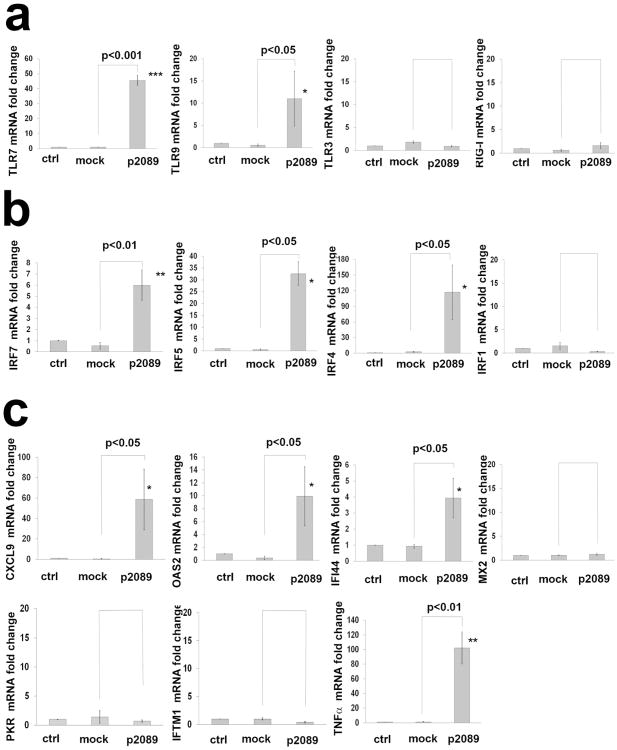
To explore the interaction between EBV and fibroblast innate immune responses, we examined the expression of TLR activated-genes in infected-cells at 4 weeks PI. Expression of TLR7 and TLR9 mRNA was significantly induced in EBV/p2089-infected-SSc-fibroblasts (Fig. 3a), as well as the mRNA levels of IRF7, IRF5 and IRF4 and selected Interferon-stimulated-genes (ISGs) (Fig. 3a-c). Remarkably, TNFα, a gene found modulated by EBV in B-lymphocytes, was also robustly increased in EBV/p2089-infected-SSc-fibroblasts. No increased expression of TLRs, IRFs or ISGs were observed in mock-infected- or in parallel uninfected-fibroblast-cultures.
Figure 3. EBV activates innate antiviral response in infected-SSc-fibroblasts.
Monocytes (MO), Dendritic Cells (DCs) from healthy donors (HDs) bound to EBV-p2089 were co-cultured with dermal fibroblasts from SSc patients. MO and DCs not exposed to EBV-p2089 were co-cultured with SSc fibroblasts as mock-infection; fibroblasts left untreated were used as control. After 72h MO, DCs and EBV-p2089 free virus were removed from fibroblasts cultures and total RNA extracted after 4 weeks post infection. (a-c) mRNA expression of TLRs, IRFs, ISGs and TNF in EBVp2089-infected, mock-infected and control fibroblasts from SSc patients, evaluated by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 5 separate SSc-fibroblast-lines. p-values calculated using two-tailed T-test.
EBV induces a profibrotic phenotype in infected-SSc-fibroblasts
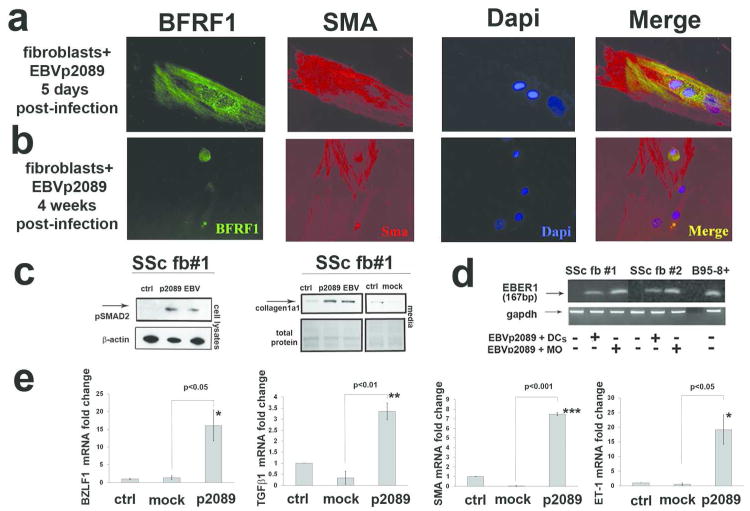
Next we asked whether viral interaction with SSc-fibroblasts might also promote proliferation and expression of activation markers as it does in B-cells (Miller et al., 1972). Specifically, we evaluated expression of genes and protein known to be related to SSc pro-fibrotic-phenotype (Varga and Abraham, 2007). We found co-expression of EBV lytic-protein BFRF1 and SMA-antigen in SSc-fibroblasts, and increased expression of genes implicated in fibroblast-myofibroblast conversion such as TGFβ1, EDN1 and SMA, and several TGFβ-regulated genes (EGR1, PAI1, COMP and COLIV) in EBV/p2089-infected-SSc-fibroblasts at 4 weeks PI (Fig. 4a-b,e and Fig. S7). A persistent activation of phospho-SMAD2, a critical mediator of TGFβ induced collagen secretion, was also detected in the cell-lysates of SSc-fibroblasts infected either by EBV/p2089 or EBV (Fig. 4c).
Figure 4. EBV induces myofibroblasts activation markers in infected-SSc-fibroblasts.
(a-b) Double IF of adherent fibroblasts stained with EBV-early/lytic-BFRF1 and αSMA-antigen (red) as indicated. Diaminidino-2-phenylindole (DAPI) was used as counterstaining for the nuclei (scale bar 50 μm). (c) Western-blot was performed to determine pSMAD2 and type I collagen secretion in cell-lysates and in the media of indicated fibroblasts cultures at 4-week-post-infection (PI). Total protein loading was determined by Ponceau-S staining. (d) Representative PCR products of EBV-DNA in 2 SSc-fibroblast-cell-lines infected with Monocytes (MO)-EBV-p2089 and dendritic-cells (DCs)-EBV-p2089 after 4 week PI; DNA from B95-8-EBV-positive cells were used as positive control. (e) mRNA expression of indicated genes in EBV-p2089-infected and in mock-infected-fibroblasts 4-week PI, evaluated by q-PCR. Bars represent mean ± S.E.M. from 5 separate SSc-fibroblast-lines. p-values calculated using two-tailed T-test
TLR7 and TLR9 agonists stimulate IFN-regulated genes in fibroblasts from healthy control skin
To evaluate whether activation of TLR7/TLR9 mimics the innate immune modulation induced by EBV in infected-SSc-fibroblasts, parallel fibroblast cultures generated from the same SSc patients and HDs not exposed to the virus, were stimulated with TLR-ligands, namely R837/imiquimod (for TLR7) and CpGODN2006 oligonucleotide (for TLR9) (Paun et al., 2008; Schoenemeyer et al., 2005).
We found that activation of the TLR-pathway by CpGODN2006, or a combination of R837 and CpGODN2006 ligands, significantly induced MX2, OAS2 and CXCL9 expression in HD-fibroblasts 24h after treatment (Fig. S8a); chronic activation of TLR-pathway induced CXCL9 expression in HD-fibroblasts treated for 4 weeks (Fig. S9a). No expression of TGFβ-regulated genes and/or increase of collagen secretion were detected in acute/chronic TLR-stimulated HD-fibroblasts (Fig. S8c and data not shown). In contrast, we did not find activation of IFN-inducible-gene-responses, as well as any increase of collagen secretion upon acute/chronic stimulation by TLR7/9-ligand-agonists in SSc-fibroblasts (Fig. S8b,d and Fig. S9b).
Discussion
We show here that EBV DNA, mRNAs and proteins are present in SSc skin, and the majority of the cells expressing EBER RNA are fibroblasts. To investigate the consequences of EBV infection in stromal cells we developed an innovative methodology that requires MO and DCs as a transfer vehicle for infection of fibroblasts. Here, we report that EBV is able to infect human fibroblasts “in vitro” and activate a fibroblast innate immune response. Importantly, EBV infection of SSc-fibroblasts promotes the SSc-profibrotic-phenotype, inducing an aberrant TLR activation pathway that is responsible for expression of IRFs, selected-ISGs, TGFβ1 and several markers of fibroblast activation, such as SMA and Endothelin-1 in infected-SSc-cells. Our study provides mechanistic insight into how EBV infection of stromal cells impacts fibroblast-myofibroblast conversion, which is consistent with the phenotype seen in SSc skin.
EBV infection in LdSSc and in NLdSSc skin
We detected expression of the EBV transactivator-BZLF1-gene, latency genes, and early-lytic protein in vivo in PBMCs, as well as in the skin, indicating that EBV infection is systemic and ongoing in SSc. Intriguingly, EBV expression of EBER, BZLF1 and EBNA1, and EBV proteins for ZEBRA, BFRF1 and BFLF2 were expressed in LdSSc and prominently in NLdSSc, suggesting that a similar “EBV footprint” was found in areas not associated with fibrosis. Previous studies have shown almost identical disease-specific-patterns of gene expression from LdSSc, NLdSSc skin biopsies and from fibroblast cultures, revealing that both LdSSc and NLdSSc shared the same gene expression (Fuzii et al., 2008; Whitfield et al., 2003); our finding of EBV transcriptional programs in SSc skin are in line with these data. Since the mechanism of how the virus persists in certain infected cells and not in others it is still not clear, a possible explanation would be that the innate immune system of infected cells does not allow EBV to survive in the context of NLdSSc-skin.
Moreover, in SSc skin biopsies we found that SMCs, myo-epithelial cells and most endothelial cells expressed EBER RNA, suggesting that EBV tropism in vivo could be broader than initially thought and extended to other cell types as permissive targets in SSc.
EBV and Innate immunity
EBV has long been proposed as a common associated factor for autoimmune diseases: SLE, Rheumatoid arthritis (RA) and multiple sclerosis (MS), since EBV latent-antigens and high titers of EBV antibodies have been detected more often in these patients; moreover high titers of IgM-VCA-antibodies were found in SSc, implying a recent EBV infection in this disease (Arnson et al., 2009; Lunemann and Munz, 2007; Niller et al., 2008). In support of this association, LMP1-antigen mRNA was detected in SSc skin, however the cellular source of the EBV product was not identified (Vaughan et al., 2000). We show here that EBV-early-lytic infection occurs in fibroblasts of SSc patients, suggesting that fibroblasts might represent the crucial target of EBV-infection in SSc skin.
Several mechanisms have been described to explain how EBV triggers autoimmune disease, such as antigen cross-reactivity with self-nuclei protein and/or bystander activation of autoreactive cells (Iwakiri et al., 2009; Langland et al., 2006; Munz et al., 2009; Samanta et al., 2008). In this study we reveal an unreported feature employed by EBV that involves viral RNA and DNA triggering the innate immune system of non-immune cells. Specifically we show that EBV triggers the innate immune system activating TLR-like antiviral responses. The contribution of EBV infection to the innate immune system is unexplored in fibroblasts. EBV activates TLR7 in B-cells although additional mechanisms have recently been proposed to explain how EBV might activate TLRs or different pattern-recognition-receptors in other cellular systems (Lindhout et al., 1994; Martin et al., 2007; Samanta et al., 2008). We found that EBV increases mRNA levels of IRF5/IRF7 and TLR7/9, suggesting that EBV might signal through the TLR7/9-pathway in infected-fibroblasts, however further experiments are required to address this issue. Several studies showed that IRF5 and IRF7 are critical mediators of TLR7/TLR9 signaling in response to ssRNA and CpGDNA in several cell types including fibroblasts (Schoenemeyer et al., 2005; Tamura et al., 2008). Moreover, IRF5 and IRF7 seem to be equally required by the host and the virus, since both are crucial for the host in activating the IFN-system and for the virus in regulating several EBV proteins necessary for viral B-cells transformation in infected-cells (Barton et al., 2011; Ning et al., 2003; Ning et al., 2005; Paun and Pitha, 2007; Savitsky et al., 2010).
ISGs, TGFβ and EBV
We found that activation of TLR-signaling by EBV promotes fibroblast antiviral response that culminate with the induction of selected ISGs and TGF β-regulated-genes, which have been found to be increased in PBMCs (TNFα, CXCL9, OAS2), and skin of patients with SSc (CXCL9, OAS2, SMA, COMP and EGR1) (Farina et al., 2010b; Radstake et al., 2010). In addition, we also found increased SMAD2 phosphorylation and collagen proteins in EBV-infected-SSc-fibroblasts, suggesting that EBV directly activates the TGFβ-transduction-pathway. As expected, we found that activation of TLR7/TLR9 by R837 and CpGBODN ligands significantly induced ISGs in HD-fibroblasts (MX2, IFI44, OAS2, CXCL9); nevertheless none of these ligands induced TGFβ-regulated-genes and collagen in HD-fibroblasts, suggesting that TLR-activation in the absence of chronic EBV-infection is unable to stimulate a TGFβ-response. Intriguingly, selected-ISGs such as PKR, MX2 and TLR3, which are transcriptionally regulated by type I IFN, were not induced by the virus, suggesting that EBV up-regulates a distinct “IFN signature” incompetent to fully stimulate the protective IFN-response in infected-SSc-fibroblasts. It is well accepted that EBV is a poor IFN inducer permitting efficient lytic replication in B-cells, although the likelihood that it might happen in fibroblasts is unexplored (Kikuta, 1986; Spender et al., 2001). Specifically EBV has evolved multiple strategies to evade the immune system specifically suppressing and/or blocking several pathways of the IFN-response, mainly by inhibiting IRF7 activity (Bentz et al., 2010; Elia et al., 1996; Hahn et al., 2005; Langland et al, 2006; Michaud et al., 2010; Samanta and Takada, 2010; Wang et al., 2009; Wang et al., 2011; Wu et al, 2009). Our data show that EBV induces expression of IRF4 in infected fibroblasts, suggesting that it might serve as repressor of IFNα/β in fibroblasts as it is known in immune-cells (Hrdlickova et al., 2001; Negishi et al., 2005; Paun and Pitha, 2007; Wang et al., 2011).
EBV and fibroblasts infection
Although fibroblasts are negative for the EBV-receptor CD21, spontaneous EBV-fibroblast-infection was detected in a primary cell line from RA synovial tissue (Koide et al., 1997). We did not detect EBV-DNA in SSc-fibroblast-cell-lines (Fig. 4d). Here we report a system that successfully infects fibroblasts “in vitro”, providing evidence that EBV is able to infect human dermal fibroblasts using MO or DCs as a vehicle for infection (Lindhout et al., 1994; Savard et al., 2000). It is likely that EBV uses alternative strategies to infect fibroblasts that bypass the absence of CD21, similar to the described transmission of EBV to human epithelial cells by cell-to-cell contact (Janz et al., 2000). There is a growing interest in exosomes, the specialized membranous vesicles derived from the endocytic compartment that can carry and deliver functional mRNA, miRNAs and proteins to various cells (Pegtel et al., 2010; Valadi et al., 2007; Zomer et al., 2010). Thousands of EBV-miRNA copy-numbers have been detected in exosomes from LCL infected-cells, suggesting that EBV-containing exosomes may be continuously secreted and transferred from the infected-cells to uninfected neighboring cells (Pegtel et al., 2010; Wurdinger et al., 2012). Possibly EBV-infected-immune-cells might transfer functional EBV-RNA and protein to fibroblasts through exosomes.
Our data show that EBV persists in fibroblasts exploiting both lytic- and latent-cycles. We did not detect any late-viral-product-gp350/220 and/or mature virions in SSc biopsies, suggesting that EBV replication is not complete in infected-SSc-fibroblasts and possibly occurs in an abortive-cycle, whereas viral DNA might be conserved in growing fibroblast cultures. Previous studies showed that the EBV lytic-cycle is abortive in several EBV-associated diseases and in specific infected-cells (Al Tabaa et al., 2009; Al Tabaa et al., 2011; Bibeau et al., 1994; Martel-Renoir et al., 1995); perhaps fibroblasts do not provide the necessary environment for productive infection so that the virus is not able to perform the normal replication program.
EBV/p2089-recombinant and EBV/B95-8-cell-derived-virus show similar cellular tropism in infecting LdSSc, NLdSSc and HD-fibroblasts “in vitro”, suggesting that EBV-infection can occur in mesenchymal cells. Intriguingly, we noticed that LdSSc and to a lesser degree NLdSSc as well as HD-fibroblast-lines could all be infected by EBV, however only fibroblasts from LdSSc and occasionally from NLdSSc-skin were able to support sustained viral presence for up to 6 months. These results suggest that the LdSSc-fibroblast phenotype might predispose to EBV-chronic-infection, promoting EBV survival in the cells, whereas fibroblasts from HDs appear to be able to control EBV infection, clearing the virus. Possibly EBV-infection spontaneously resolves in the context of presumably immunocompetent conditions, where infected-cells, in our case from HDs, might still have a competent immune system able to induce a full IFNs-response controlling the EBV-infection. Characterization of “EBV-IFN-signature” deserves further investigation specifically in these cells.
EBV and SSc-profibrotic-phenotype
Activated fibroblasts are considered the principal mediators of fibrogenesis in SSc. It is known that SSc fibroblasts show a profibrotic phenotype with sustained TGFβ-activation, increased collagen production, and increased number of myofibroblasts (Varga and Abraham, 2007). We found that the EBV-lytic cascade is associated with upregulation of TGFβ1, several TGFβ-regulated genes, including SMA, and increased collagen by infected fibroblasts. Intriguingly, the EBV transactivator-BZLF1-gene has been linked to development of fibrosis in some other EBV-associated diseases, although its primary role is to disrupt viral latency and transactivate the expression of early-lytic genes (Grogan et al., 1987; Guenther et al., 2010). Specifically BZLF1 was shown to interact with numerous key cellular transcriptional regulatory factors including TGFβ1/3 and EGR1 in infected-epithelial-cells (Adamson and Kenney, 1999; Cayrol and Flemington, 1995; Chang et al., 2006; Tsai et al., 2009). Thus BZLF1 interaction with one of these factors might be also responsible for SMAD2 phosphorylation and TGFβ1 increased expression in EBV-infected-SSc-fibroblasts.
Previous studies have suggested a role for EBV-infection as in the pathogenesis of smooth muscle tumors (SMT) in patients with clinical immunosuppression. Specifically EBER was found expressed in SMT-cells and in myofibroblasts from sclerosing-nodular-trasformation of the spleen, suggesting that EBV-infected-myofibroblasts could be a common pathway of a fibrosclerotic process occurring in splenic inflammatory tumor-like lesion, although the origin of EBER+myofibroblasts was not fully understood (Kashiwagi et al., 2008; Lee et al., 1995; Weinreb et al., 2007). Our in vitro data showed that EBV induces a myofibroblast phenotype in infected-SSc-fibroblasts.
Our results show that SSc-fibroblasts have greatly diminished IFN-inducible-gene response upon TLR7/9-agonist stimulation, possibly one of the reasons that EBV is able to infect and persist in SSc-fibroblasts. Further experiments are needed to understand whether EBV-infection might induce epigenetic changes in infected-fibroblasts, as it does in B-, Lymphoblastoid- and epithelial-cells, blunting the IFN response in “transformed” cells (Banerjee et al., 2013; Gregorovic et al., 2011; Queen et al., 2013).
Overall our study provides compelling evidence that EBV infection could contribute to fibrosis in SSc skin through multiple factors and a combination of subsequent pathological events such as virus-host-cellular-interactions that may lead to aberrant activation of TLR-pathway in infected-SSc-fibroblasts. This pathway activates selected ISGs, cytokines, and influences fibroblast-pro-fibrotic-phenotype driving myofibroblast-conversion. These results indicate that EBV-infection might cause the patho-immunogenetic-alterations seen in SSc-fibroblasts, and those abnormalities are related to the viral-strategies that subvert the host-innate-immune response in infected-cells.
Concurrent EBV infection occurring in mesenchymal, endothelial, and immune cells of SSc patients may underlay the main pathological features of SSc including autoimmunity, vasculopathy and fibrosis, and provide a unified disease mechanism represented by EBV reactivation.
Methods
Study subjects
All study subjects met the criteria for SSc as defined previously (LeRoy et al., 1988). The study was conducted under a protocol in adherence to the Helsinki Guidelines and approved by the Boston University Medical Center, Institutional Review Board and all subjects gave written informed consent. Skin biopsies were performed as previously described (Farina et al., 2010a; Farina et al., 2010b)
EBER “in situ” hybridization
In situ hybridization (ISH) for EBV encoded RNA (EBER) was carried out using fluorescin isothiocynate (FITC-l-labeled-peptide-nucleic-acid probed for EBER and peptide-nucleic-acid -ISH detection kit (DAKO, Carpinteria, CA) on paraffin-embedded skin sections according to the manufacturer's protocol.
Virus preparation
EBV was obtained from producing-B95.8 cell-line and from p2089-cell-line as previously described (Delecluse et al., 1998; Farina et al., 2000).
Isolation of Monocytes and monocyte-derived DC from HDs
PBMCs from 10 HDs were isolated by Ficoll-Paque-gradient-centrifugation (Pharmacia, Uppsala, Sweden). CD14+/monocytes were positively selected using anti-CD14-MAb-conjugated-magnetic-microbeads (MiltenyiBiotec, Auburn, CA) confirmed by flow cytometry (Cirone et al., 2007). Each group of infection (n=6) were carried out using fibroblasts from SSc and HDs, and MO/DCs from a single HD at the time. MO/DCs isolated from different HDs were used to infect the same SSc-fibroblast-cell-lines. CD14+/- and BDCA1+/- selection markers were also evaluated by qPCR.
Transfer-infection
Donor cells (MO and DCs) obtained from 10 HDs were exposed to EBV- and/or -p2089 at known multiplicities of infection for 3 h at 4°C; after extensive washing 104/ cells were added to confluent fibroblasts culture in 8/well-chamber-slides (Feederle et al., 2007). 72h after co-cultivation with human primary fibroblast, supernatants (MO/DCs) were removed and cultured separately from the infected fibroblast. After 2-days, total RNA was extracted from MO and DCs. Transfer-infection in fibroblasts cultures was assayed 72h after the initiation of co-culture by counting the percentage of GFP-positive-cells in trypsinized acceptor cell-suspensions. p2089-infected- mock-infected and control-fibroblasts were cultured in D-MEM (10%FCS) and monitored by IFA or PCR up to 6 months post infection. Total RNA was extracted from p2089-infected-, mock-infected and control-SSc-fibroblasts 4 weeks PI.
Statistical analyses
All data are expressed as the mean±S.E. The means between two groups were analyzed by Student's t test, Wilcoxon test and Fisher-exact-test. Significance was taken at P<0.05.
Supplementary Material
Fig. S1. EBER probe in the skin of healthy donors.
(a and b) Representative images of EBER in situ hybridization (ISH) in the skin from 2 healthy donors (HDs) (left panels); α-smooth muscle actin (αSMA) staining by IHC in serial sections from skin of 2 HDs (right-panels) (scale/bars upper-panels, 100μm, lower-panel 50μm).
Fig. S2. Expression of EBV transcripts in skin and PBMCs of SSc patients.
(a) Representative gel electrophoresis of EBV-lytic/latency genes (BZLF1/EBNA1) RT-PCR products from 6 lesional diffuse SSc (LdSSc) and one representative normal skin (HD); 293 and Raji cells were use as negative and positive control respectively. (b) RT-PCR products of EBV-lytic-gene BZLF1 in (LdSSc) and non/lesional (NLdSSc) from 2 representative patients. (c) RT-PCR products of EBV-lytic-gene BZLF1 in PBMCs from dSSc and 2 representative HDs subjects. (d) PCR products of EBV DNA (EBER1) in LdSSc and in 2 HDs representative skin; DNA from B95-8-EBV-positive cells were used as positive control, GAPDH used as internal control; incidence of EBV DNA and BZLF1/EBNA1 transcripts in screened skin section are summarized in Supplemental Table 2. (e-f) Detection of EBV-load by q-PCR; each sample was tested in duplicates and normalized by endogenous internal control. Shown here are copies of viral nucleic acid in the skin calculated by standard curve. 293 cells and Foreskin dermal skin were used as negative control. The average of copies number is represented by horizontal line ± SE. p-values calculated using Wilcoxon two samples test.
Fig. S3. Expression of EBV proteins in the skin of SSc patients.
(a-c) Immuno-histochemistry (IHC) in serial tissue sections from lesional (LdSSc) and non-lesional (NLdSSc) skin samples. Nuclear localization of EBV-lytic-protein/Zebra in scattered fibroblasts (square insert) and in the matrix, and expression of the early lytic BFLF2 protein in LdSSc deep dermis obtained from the same patient (a-b). Expression of Zebra and early-lytic-BFLF2 proteins in LdSSc and NLdSSc skin from the same patient (c). Crude lysate from skin and PBMCs of representative SSc patients and HDs (d and e), skin and PBMCs samples obtained from the same patients (f), were separated on SDS-PAGE, blotted on PVDF and probed with the indicated antibodies; lysate from B95-8 EBV infected cells were used as positive control; β-actin was used as loading control. Numbers represent distinct patients enrolled in the study. (red-staining bar scale 100μm (upper-panels) and 10μm (lower and squares panels).
Fig. S4. EBV RNAs and antigens in the skin of SSc patients.
(a-c) Representative images of EBERs in situ hybridization (ISH), and (d-f) immuno-histochemistry (IHC) of EBV-lytic-protein Zebra in serial tissue sections from lesional skin (LdSSc) of two SSc cohort patients naïve vs immunosuppressed treatment (arrows indicate vessels positive or negative for EBERs staining). Numbers on the side represent distinct patient enrolled in the study, whose clinical characteristics are summarized in Table S5. Bar scale 100μm (upper panels) and 10μm (lower and squares panels)
Fig. S5. EBV antigens expression in the skin.
Immuno-histochemistry (IHC) in serial skin tissue sections from lesional (LdSSc), non-lesional (NLdSSc) and healthy-donor (HD) skin sample of indicated EBV-proteins (red-staining bar scale 50μm). Numbers on the side represent distinct patient enrolled in the study.
Fig. S6. EBV-p2089-recombinant-virus infects human SSc-fibroblasts “in vitro”.
(a) Inverted microscope image of 2 EBV-p2089-infected-SSc-fibroblast 4 week/post infection (PI). GFP expression indicates recombinant ebv- infected cells. (left panel: phase-contrast-light-microcopy; bar scale 20 mm). (b) Western-blot analysis of Poly (ADP-ribose) polymerase (PARP) protein in cell lysates from EBV-p2089 and mock-infected-SSc-fibroblast-cultures at 4-week-PI. B95-8 EBV-activated was used as positive control.
Fig. S7 Expression of TGFβ-responsive genes in fibroblasts infected with EBV.
mRNA expression of indicated genes in EBVp2089-infected, mock-infected and control fibroblasts from SSc patients after 4/week post infection, evaluated by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 3 separate experiments from different SSc-fibroblast-cell-lines. p-values calculated using two-tailed T-test. *= p<0.05; **= p<0.01; ***= p<0.001
Fig. S8. Expression of Interferon-stimulated-genes (ISGs) in human dermal fibroblasts by TLRs stimulation.
Fibroblasts explanted from healthy donors (HDs) (a) and from lesional skin of patients with dSSc (b), were starved o/n and incubated with TLR-agonist-ligands as indicated for 24hrs. mRNA was harvested and analyzed by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 3 separate experiments using 3 different cell lines. p-values calculated using two-tailed T-test. *= p<0.05; **= p<0.01; ***= p<0.001. (c-d) Western blot analysis was performed to determine type I collagen secretion in the media of indicated fibroblasts cultures after indicated treatment. Total protein loading was determined by Ponceau-S staining of the filter after western transfer (bottom panel).
Fig. S9.Expression of Interferon-stimulated-genes (ISGs) in human dermal fibroblasts by TLRs chronic stimulation.
Fibroblasts explanted from healthy donors (HDs) (a) and from lesional skin of patients with dSSc (b), were incubated with TLR-agonist-ligands as indicated and treated for 3 times/week for 4 weeks in presence of FBS 10%. mRNA was harvested and analyzed by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 3 separate experiments using 3 different cell lines. p-values calculated using two-tailed T-test. *= p<0.05; **= p<0.01; ***= p<0.001.
Table S1. Demographics and Clinical characteristics of SSc patients and Healthy Donor (HD) subjects
Table S2. Detection of Epstein-Barr virus genomes and their products in PBMCs and skin tissues of patients with SSc and HD subjects
1: DNA detected by PCR
Table S3: serological profile to EBV in patients with diffuse (dSSc) and limited (lSSc) SSc disease.
Anti-VCA* = antibody to the viral capsid antigen, positive >1.1 U/mL; negative < 0.37;
n/a**= not available
Table S4. Demographics and clinic characteristics of two cohorts of SSc patients Treatment Naïve vs Treatment
LdSSc = Lesional diffuse SSc
*d.d. mo= duration of disease in months; MRSS: Modified Rodnan skin score
Table S5. Comparison of EBER “in situ” RNA expression in SSc patients Treatment-Naïve vs Treatment-Immunosuppressed
Table S6. (part I): EBV expression pattern in lesional skin (LdSSc) and auto-antibodies profile in patients with SSc diffuse disease.
Table S6. (part II): EBV expression pattern in lesional skin (lSSc) and auto-antibodies profile in patients with limited SSc disease.
Table S7. Detection of Epstein-Barr virus products in PBMCs and skin tissues of patients with SSc and HDs
*= evaluated by Western-blot
Table S8. Real-time PCR primers used for detection of viral cDNAs.
Acknowledgments
We wish to thank Dr. Henri-Jacques Delecluse, Dr. Russell Widom and Dr. Jeff Browning for helpful advices and critical reading of the manuscript and Claudia Zompetta for technical support. This manuscript is dedicated to the memory of Dr. Joseph H. Korn. This study was supported by: NIH-NIAMS grant 1R03AR062721-01 and “Norma Nadeau/Mary-Van-Neste-Research Grant” New England chapter of the Scleroderma Foundation (to G.A. Farina); Associazione Italiana per la ricerca sul Cancro (to A.Faggioni); NIH-NIAMS grant 5P500AR060780-02, 5P30AR061271-02 and 5R1AR1089-07 (to R.Lafyatis).
Footnotes
The authors have no conflicting financial interests
Conflict of Interest: The authors state no conflict of interest.
AF and GAF designed experiments; AF, GAF, MC, MY, SL, CP, SM performed experiments; GAF, AFag and RL provided reagents; AF, AFag, MT, RL and GAF prepared the manuscript
Supplementary Material: Refer to Web version on PubMed Central for supplementary material
References
- Adamson AL, Kenney S. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. Journal of virology. 1999;73:6551–8. doi: 10.1128/jvi.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Tabaa Y, Tuaillon E, Bollore K, Foulongne V, Petitjean G, Seigneurin JM, et al. Functional Epstein-Barr virus reservoir in plasma cells derived from infected peripheral blood memory B cells. Blood. 2009;113:604–11. doi: 10.1182/blood-2008-02-136903. [DOI] [PubMed] [Google Scholar]
- Al Tabaa Y, Tuaillon E, Jeziorski E, Ouedraogo DE, Bollore K, Rubbo PA, et al. B-cell polyclonal activation and Epstein-Barr viral abortive lytic cycle are two key features in acute infectious mononucleosis. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2011;52:33–7. doi: 10.1016/j.jcv.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Arnson Y, Amital H, Guiducci S, Matucci-Cerinic M, Valentini G, Barzilai O, et al. The role of infections in the immunopathogensis of systemic sclerosis--evidence from serological studies. Annals of the New York Academy of Sciences. 2009;1173:627–32. doi: 10.1111/j.1749-6632.2009.04808.x. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Lu J, Cai Q, Saha A, Jha HC, Dzeng RK, et al. The EBV Latent Antigen 3C Inhibits Apoptosis through Targeted Regulation of Interferon Regulatory Factors 4 and 8. PLoS pathogens. 2013;9:e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E, Mandal P, Speck SH. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annual review of immunology. 2011;29:351–97. doi: 10.1146/annurev-immunol-072710-081639. [DOI] [PubMed] [Google Scholar]
- Bentz GL, Liu R, Hahn AM, Shackelford J, Pagano JS. Epstein-Barr virus BRLF1 inhibits transcription of IRF3 and IRF7 and suppresses induction of interferon-beta. Virology. 2010;402:121–8. doi: 10.1016/j.virol.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibeau F, Brousset P, Knecht H, Meggetto F, Drouet E, Rubin B, et al. Epstein-Barr virus replication in Hodgkin disease. Bulletin du cancer. 1994;81:114–8. [PubMed] [Google Scholar]
- Cayrol C, Flemington EK. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor beta igh3 (TGF-beta igh3) and TGF-beta 1. Journal of virology. 1995;69:4206–12. doi: 10.1128/jvi.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Lee HH, Chen YT, Lu J, Wu SY, Chen CW, et al. Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. Journal of virology. 2006;80:7748–55. doi: 10.1128/JVI.02608-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirone M, Lucania G, Bergamo P, Trivedi P, Frati L, Faggioni A. Human herpesvirus 8 (HHV-8) inhibits monocyte differentiation into dendritic cells and impairs their immunostimulatory activity. Immunology letters. 2007;113:40–6. doi: 10.1016/j.imlet.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8245–50. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus DH. Autoimmune disease: A role for new anti-viral therapies? Autoimmunity reviews. 2011;11:88–97. doi: 10.1016/j.autrev.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Ebrahimi B, Dutia BM, Brownstein DG, Nash AA. Murine gammaherpesvirus-68 infection causes multi-organ fibrosis and alters leukocyte trafficking in interferon-gamma receptor knockout mice. The American journal of pathology. 2001;158:2117–25. doi: 10.1016/s0002-9440(10)64683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A, Laing KG, Schofield A, Tilleray VJ, Clemens MJ. Regulation of the double-stranded RNA-dependent protein kinase PKR by RNAs encoded by a repeated sequence in the Epstein-Barr virus genome. Nucleic acids research. 1996;24:4471–8. doi: 10.1093/nar/24.22.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Santarelli R, Gonnella R, Bei R, Muraro R, Cardinali G, et al. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. Journal of virology. 2000;74:3235–44. doi: 10.1128/jvi.74.7.3235-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina G, Lafyatis D, Lemaire R, Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis and rheumatism. 2010a;62:580–8. doi: 10.1002/art.27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina G, York M, Collins C, Lafyatis R. dsRNA activation of endothelin-1 and markers of vascular activation in endothelial cells and fibroblasts. Annals of the rheumatic diseases. 2011;70:544–50. doi: 10.1136/ard.2010.132464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina GA, York MR, Di Marzio M, Collins CA, Meller S, Homey B, et al. Poly(I:C) drives type I IFN- and TGFbeta-mediated inflammation and dermal fibrosis simulating altered gene expression in systemic sclerosis. The Journal of investigative dermatology. 2010b;130:2583–93. doi: 10.1038/jid.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle R, Neuhierl B, Bannert H, Geletneky K, Shannon-Lowe C, Delecluse HJ. Epstein-Barr virus B95.8 produced in 293 cells shows marked tropism for differentiated primary epithelial cells and reveals interindividual variation in susceptibility to viral infection. International journal of cancer Journal international du cancer. 2007;121:588–94. doi: 10.1002/ijc.22727. [DOI] [PubMed] [Google Scholar]
- Fuzii HT, Yoshikawa GT, Junta CM, Sandrin-Garcia P, Fachin AL, Sakamoto-Hojo ET, et al. Affected and non-affected skin fibroblasts from systemic sclerosis patients share a gene expression profile deviated from the one observed in healthy individuals. Clinical and experimental rheumatology. 2008;26:866–74. [PubMed] [Google Scholar]
- Green M, Bueno J, Rowe D, Mazariegos G, Qu L, Abu-Almagd K, et al. Predictive negative value of persistent low Epstein-Barr virus viral load after intestinal transplantation in children. Transplantation. 2000;70:593–6. doi: 10.1097/00007890-200008270-00010. [DOI] [PubMed] [Google Scholar]
- Gregorovic G, Bosshard R, Karstegl CE, White RE, Pattle S, Chiang AK, et al. Cellular gene expression that correlates with EBER expression in Epstein-Barr Virus-infected lymphoblastoid cell lines. Journal of virology. 2011;85:3535–45. doi: 10.1128/JVI.02086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan E, Jenson H, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:1332–6. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther JF, Cameron JE, Nguyen HT, Wang Y, Sullivan DE, Shan B, et al. Modulation of lung inflammation by the Epstein-Barr virus protein Zta. American journal of physiology Lung cellular and molecular physiology. 2010;299:L771–84. doi: 10.1152/ajplung.00408.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn AM, Huye LE, Ning S, Webster-Cyriaque J, Pagano JS. Interferon regulatory factor 7 is negatively regulated by the Epstein-Barr virus immediate-early gene, BZLF-1. Journal of virology. 2005;79:10040–52. doi: 10.1128/JVI.79.15.10040-10052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova R, Nehyba J, Bose HR., Jr Interferon regulatory factor 4 contributes to transformation of v-Rel-expressing fibroblasts. Molecular and cellular biology. 2001;21:6369–86. doi: 10.1128/MCB.21.19.6369-6386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D, Zhou L, Samanta M, Matsumoto M, Ebihara T, Seya T, et al. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. The Journal of experimental medicine. 2009;206:2091–9. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz A, Oezel M, Kurzeder C, Mautner J, Pich D, Kost M, et al. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. Journal of virology. 2000;74:10142–52. doi: 10.1128/jvi.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi S, Kumasaka T, Bunsei N, Fukumura Y, Yamasaki S, Abe K, et al. Detection of Epstein-Barr virus-encoded small RNA-expressed myofibroblasts and IgG4-producing plasma cells in sclerosing angiomatoid nodular transformation of the spleen. Virchows Archiv : an international journal of pathology. 2008;453:275–82. doi: 10.1007/s00428-008-0648-z. [DOI] [PubMed] [Google Scholar]
- Kikuta H. IFN production by Epstein-Barr virus in human mononuclear leukocytes and suppression of the virus-induced transformation by the endogenous interferon. [Hokkaido igaku zasshi] The Hokkaido journal of medical science. 1986;61:46–57. [PubMed] [Google Scholar]
- Koide J, Takada K, Sugiura M, Sekine H, Ito T, Saito K, et al. Spontaneous establishment of an Epstein-Barr virus-infected fibroblast line from the synovial tissue of a rheumatoid arthritis patient. Journal of virology. 1997;71:2478–81. doi: 10.1128/jvi.71.3.2478-2481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus research. 2006;119:100–10. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Larsen M, Sauce D, Deback C, Arnaud L, Mathian A, Miyara M, et al. Exhausted cytotoxic control of Epstein-Barr virus in human lupus. PLoS pathogens. 2011;7:e1002328. doi: 10.1371/journal.ppat.1002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Locker J, Nalesnik M, Reyes J, Jaffe R, Alashari M, et al. The association of Epstein-Barr virus with smooth-muscle tumors occurring after organ transplantation. The New England journal of medicine. 1995;332:19–25. doi: 10.1056/NEJM199501053320104. [DOI] [PubMed] [Google Scholar]
- LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. The Journal of rheumatology. 1988;15:202–5. [PubMed] [Google Scholar]
- Lindhout E, Lakeman A, Mevissen ML, de Groot C. Functionally active Epstein-Barr virus-transformed follicular dendritic cell-like cell lines. The Journal of experimental medicine. 1994;179:1173–84. doi: 10.1084/jem.179.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunemann JD, Munz C. Epstein-Barr virus and multiple sclerosis. Current neurology and neuroscience reports. 2007;7:253–8. doi: 10.1007/s11910-007-0038-y. [DOI] [PubMed] [Google Scholar]
- Martel-Renoir D, Grunewald V, Touitou R, Schwaab G, Joab I. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. The Journal of general virology. 1995;76(Pt 6):1401–8. doi: 10.1099/0022-1317-76-6-1401. [DOI] [PubMed] [Google Scholar]
- Martin HJ, Lee JM, Walls D, Hayward SD. Manipulation of the toll-like receptor 7 signaling pathway by Epstein-Barr virus. Journal of virology. 2007;81:9748–58. doi: 10.1128/JVI.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorelli D, Muraro E, Merlo A, Turrini R, Fae DA, Rosato A, et al. Exploiting the interplay between innate and adaptive immunity to improve immunotherapeutic strategies for Epstein-Barr-virus-driven disorders. Clinical & developmental immunology. 2012;2012:931952. doi: 10.1155/2012/931952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud F, Coulombe F, Gaudreault E, Paquet-Bouchard C, Rola-Pleszczynski M, Gosselin J. Epstein-Barr virus interferes with the amplification of IFNalpha secretion by activating suppressor of cytokine signaling 3 in primary human monocytes. PloS one. 2010;5:e11908. doi: 10.1371/journal.pone.0011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:383–7. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz C, Lunemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nature reviews Immunology. 2009;9:246–58. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, et al. Negative regulation of Toll-like-receptor signaling by IRF-4. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15989–94. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niller HH, Wolf H, Minarovits J. Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases. Autoimmunity. 2008;41:298–328. doi: 10.1080/08916930802024772. [DOI] [PubMed] [Google Scholar]
- Ning S, Hahn AM, Huye LE, Pagano JS. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. Journal of virology. 2003;77:9359–68. doi: 10.1128/JVI.77.17.9359-9368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S, Huye LE, Pagano JS. Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling. The Journal of biological chemistry. 2005;280:12262–70. doi: 10.1074/jbc.M404260200. [DOI] [PubMed] [Google Scholar]
- Okano M, Kawa K, Kimura H, Yachie A, Wakiguchi H, Maeda A, et al. Proposed guidelines for diagnosing chronic active Epstein-Barr virus infection. American journal of hematology. 2005;80:64–9. doi: 10.1002/ajh.20398. [DOI] [PubMed] [Google Scholar]
- Pandey JP, LeRoy EC. Human cytomegalovirus and the vasculopathies of autoimmune diseases (especially scleroderma), allograft rejection, and coronary restenosis. Arthritis and rheumatism. 1998;41:10–5. doi: 10.1002/1529-0131(199801)41:1<10::AID-ART2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–53. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun A, Reinert JT, Jiang Z, Medin C, Balkhi MY, Fitzgerald KA, et al. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. The Journal of biological chemistry. 2008;283:14295–308. doi: 10.1074/jbc.M800501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen KJ, Shi M, Zhang F, Cvek U, Scott RS. Epstein-Barr virus-induced epigenetic alterations following transient infection. International journal of cancer Journal international du cancer. 2013;132:2076–86. doi: 10.1002/ijc.27893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nature genetics. 2010;42:426–9. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M, Iwakiri D, Takada K. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene. 2008;27:4150–60. doi: 10.1038/onc.2008.75. [DOI] [PubMed] [Google Scholar]
- Samanta M, Takada K. Modulation of innate immunity system by Epstein-Barr virus-encoded non-coding RNA and oncogenesis. Cancer science. 2010;101:29–35. doi: 10.1111/j.1349-7006.2009.01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard M, Belanger C, Tardif M, Gourde P, Flamand L, Gosselin J. Infection of primary human monocytes by Epstein-Barr virus. Journal of virology. 2000;74:2612–9. doi: 10.1128/jvi.74.6.2612-2619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer immunology, immunotherapy : CII. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. The Journal of biological chemistry. 2005;280:17005–12. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- Shannon-Lowe CD, Neuhierl B, Baldwin G, Rickinson AB, Delecluse HJ. Resting B cells as a transfer vehicle for Epstein-Barr virus infection of epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7065–70. doi: 10.1073/pnas.0510512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spender LC, Cornish GH, Rowland B, Kempkes B, Farrell PJ. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. Journal of virology. 2001;75:3537–46. doi: 10.1128/JVI.75.8.3537-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoolman JS, Vannella KM, Coomes SM, Wilke CA, Sisson TH, Toews GB, et al. Latent infection by gammaherpesvirus stimulates profibrotic mediator release from multiple cell types. American journal of physiology Lung cellular and molecular physiology. 2011;300:L274–85. doi: 10.1152/ajplung.00028.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annual review of immunology. 2008;26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN. TLRs and IFNs: critical pieces of the autoimmunity puzzle. The Journal of clinical investigation. 2012;122:3464–6. doi: 10.1172/JCI63835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Lin SJ, Chen PW, Luo WY, Yeh TH, Wang HW, et al. EBV Zta protein induces the expression of interleukin-13, promoting the proliferation of EBV-infected B cells and lymphoblastoid cell lines. Blood. 2009;114:109–18. doi: 10.1182/blood-2008-12-193375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Varga J, Abraham D. Systemic sclerosis: a prototypic multisystem fibrotic disorder. The Journal of clinical investigation. 2007;117:557–67. doi: 10.1172/JCI31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan JH, Shaw PX, Nguyen MD, Medsger TA, Jr, Wright TM, Metcalf JS, et al. Evidence of activation of 2 herpesviruses, Epstein-Barr virus and cytomegalovirus, in systemic sclerosis and normal skins. The Journal of rheumatology. 2000;27:821–3. [PubMed] [Google Scholar]
- Wang JT, Doong SL, Teng SC, Lee CP, Tsai CH, Chen MR. Epstein-Barr virus BGLF4 kinase suppresses the interferon regulatory factor 3 signaling pathway. Journal of virology. 2009;83:1856–69. doi: 10.1128/JVI.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Toomey NL, Diaz LA, Walker G, Ramos JC, Barber GN, et al. Oncogenic IRFs provide a survival advantage for Epstein-Barr virus- or human T-cell leukemia virus type 1-transformed cells through induction of BIC expression. Journal of virology. 2011;85:8328–37. doi: 10.1128/JVI.00570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb I, Bailey D, Battaglia D, Kennedy M, Perez-Ordonez B. CD30 and Epstein-Barr virus RNA expression in sclerosing angiomatoid nodular transformation of spleen. Virchows Archiv : an international journal of pathology. 2007;451:73–9. doi: 10.1007/s00428-007-0422-7. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Finlay DR, Murray JI, Troyanskaya OG, Chi JT, Pergamenschikov A, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12319–24. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, et al. Epstein-Barr virus LF2: an antagonist to type I interferon. Journal of virology. 2009;83:1140–6. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Gatson NN, Balaj L, Kaur B, Breakefield XO, Pegtel DM. Extracellular vesicles and their convergence with viral pathways. Advances in virology. 2012;2012:767694. doi: 10.1155/2012/767694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis and rheumatism. 2007;56:1010–20. doi: 10.1002/art.22382. [DOI] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nature reviews Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: Fit to deliver small RNA. Communicative & integrative biology. 2010;3:447–50. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. EBER probe in the skin of healthy donors.
(a and b) Representative images of EBER in situ hybridization (ISH) in the skin from 2 healthy donors (HDs) (left panels); α-smooth muscle actin (αSMA) staining by IHC in serial sections from skin of 2 HDs (right-panels) (scale/bars upper-panels, 100μm, lower-panel 50μm).
Fig. S2. Expression of EBV transcripts in skin and PBMCs of SSc patients.
(a) Representative gel electrophoresis of EBV-lytic/latency genes (BZLF1/EBNA1) RT-PCR products from 6 lesional diffuse SSc (LdSSc) and one representative normal skin (HD); 293 and Raji cells were use as negative and positive control respectively. (b) RT-PCR products of EBV-lytic-gene BZLF1 in (LdSSc) and non/lesional (NLdSSc) from 2 representative patients. (c) RT-PCR products of EBV-lytic-gene BZLF1 in PBMCs from dSSc and 2 representative HDs subjects. (d) PCR products of EBV DNA (EBER1) in LdSSc and in 2 HDs representative skin; DNA from B95-8-EBV-positive cells were used as positive control, GAPDH used as internal control; incidence of EBV DNA and BZLF1/EBNA1 transcripts in screened skin section are summarized in Supplemental Table 2. (e-f) Detection of EBV-load by q-PCR; each sample was tested in duplicates and normalized by endogenous internal control. Shown here are copies of viral nucleic acid in the skin calculated by standard curve. 293 cells and Foreskin dermal skin were used as negative control. The average of copies number is represented by horizontal line ± SE. p-values calculated using Wilcoxon two samples test.
Fig. S3. Expression of EBV proteins in the skin of SSc patients.
(a-c) Immuno-histochemistry (IHC) in serial tissue sections from lesional (LdSSc) and non-lesional (NLdSSc) skin samples. Nuclear localization of EBV-lytic-protein/Zebra in scattered fibroblasts (square insert) and in the matrix, and expression of the early lytic BFLF2 protein in LdSSc deep dermis obtained from the same patient (a-b). Expression of Zebra and early-lytic-BFLF2 proteins in LdSSc and NLdSSc skin from the same patient (c). Crude lysate from skin and PBMCs of representative SSc patients and HDs (d and e), skin and PBMCs samples obtained from the same patients (f), were separated on SDS-PAGE, blotted on PVDF and probed with the indicated antibodies; lysate from B95-8 EBV infected cells were used as positive control; β-actin was used as loading control. Numbers represent distinct patients enrolled in the study. (red-staining bar scale 100μm (upper-panels) and 10μm (lower and squares panels).
Fig. S4. EBV RNAs and antigens in the skin of SSc patients.
(a-c) Representative images of EBERs in situ hybridization (ISH), and (d-f) immuno-histochemistry (IHC) of EBV-lytic-protein Zebra in serial tissue sections from lesional skin (LdSSc) of two SSc cohort patients naïve vs immunosuppressed treatment (arrows indicate vessels positive or negative for EBERs staining). Numbers on the side represent distinct patient enrolled in the study, whose clinical characteristics are summarized in Table S5. Bar scale 100μm (upper panels) and 10μm (lower and squares panels)
Fig. S5. EBV antigens expression in the skin.
Immuno-histochemistry (IHC) in serial skin tissue sections from lesional (LdSSc), non-lesional (NLdSSc) and healthy-donor (HD) skin sample of indicated EBV-proteins (red-staining bar scale 50μm). Numbers on the side represent distinct patient enrolled in the study.
Fig. S6. EBV-p2089-recombinant-virus infects human SSc-fibroblasts “in vitro”.
(a) Inverted microscope image of 2 EBV-p2089-infected-SSc-fibroblast 4 week/post infection (PI). GFP expression indicates recombinant ebv- infected cells. (left panel: phase-contrast-light-microcopy; bar scale 20 mm). (b) Western-blot analysis of Poly (ADP-ribose) polymerase (PARP) protein in cell lysates from EBV-p2089 and mock-infected-SSc-fibroblast-cultures at 4-week-PI. B95-8 EBV-activated was used as positive control.
Fig. S7 Expression of TGFβ-responsive genes in fibroblasts infected with EBV.
mRNA expression of indicated genes in EBVp2089-infected, mock-infected and control fibroblasts from SSc patients after 4/week post infection, evaluated by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 3 separate experiments from different SSc-fibroblast-cell-lines. p-values calculated using two-tailed T-test. *= p<0.05; **= p<0.01; ***= p<0.001
Fig. S8. Expression of Interferon-stimulated-genes (ISGs) in human dermal fibroblasts by TLRs stimulation.
Fibroblasts explanted from healthy donors (HDs) (a) and from lesional skin of patients with dSSc (b), were starved o/n and incubated with TLR-agonist-ligands as indicated for 24hrs. mRNA was harvested and analyzed by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 3 separate experiments using 3 different cell lines. p-values calculated using two-tailed T-test. *= p<0.05; **= p<0.01; ***= p<0.001. (c-d) Western blot analysis was performed to determine type I collagen secretion in the media of indicated fibroblasts cultures after indicated treatment. Total protein loading was determined by Ponceau-S staining of the filter after western transfer (bottom panel).
Fig. S9.Expression of Interferon-stimulated-genes (ISGs) in human dermal fibroblasts by TLRs chronic stimulation.
Fibroblasts explanted from healthy donors (HDs) (a) and from lesional skin of patients with dSSc (b), were incubated with TLR-agonist-ligands as indicated and treated for 3 times/week for 4 weeks in presence of FBS 10%. mRNA was harvested and analyzed by qPCR. Fold-changes shown on the graph are normalized to mRNA expression by each corresponding untreated cell lines. Bars represent mean ± S.E.M. from 3 separate experiments using 3 different cell lines. p-values calculated using two-tailed T-test. *= p<0.05; **= p<0.01; ***= p<0.001.
Table S1. Demographics and Clinical characteristics of SSc patients and Healthy Donor (HD) subjects
Table S2. Detection of Epstein-Barr virus genomes and their products in PBMCs and skin tissues of patients with SSc and HD subjects
1: DNA detected by PCR
Table S3: serological profile to EBV in patients with diffuse (dSSc) and limited (lSSc) SSc disease.
Anti-VCA* = antibody to the viral capsid antigen, positive >1.1 U/mL; negative < 0.37;
n/a**= not available
Table S4. Demographics and clinic characteristics of two cohorts of SSc patients Treatment Naïve vs Treatment
LdSSc = Lesional diffuse SSc
*d.d. mo= duration of disease in months; MRSS: Modified Rodnan skin score
Table S5. Comparison of EBER “in situ” RNA expression in SSc patients Treatment-Naïve vs Treatment-Immunosuppressed
Table S6. (part I): EBV expression pattern in lesional skin (LdSSc) and auto-antibodies profile in patients with SSc diffuse disease.
Table S6. (part II): EBV expression pattern in lesional skin (lSSc) and auto-antibodies profile in patients with limited SSc disease.
Table S7. Detection of Epstein-Barr virus products in PBMCs and skin tissues of patients with SSc and HDs
*= evaluated by Western-blot
Table S8. Real-time PCR primers used for detection of viral cDNAs.