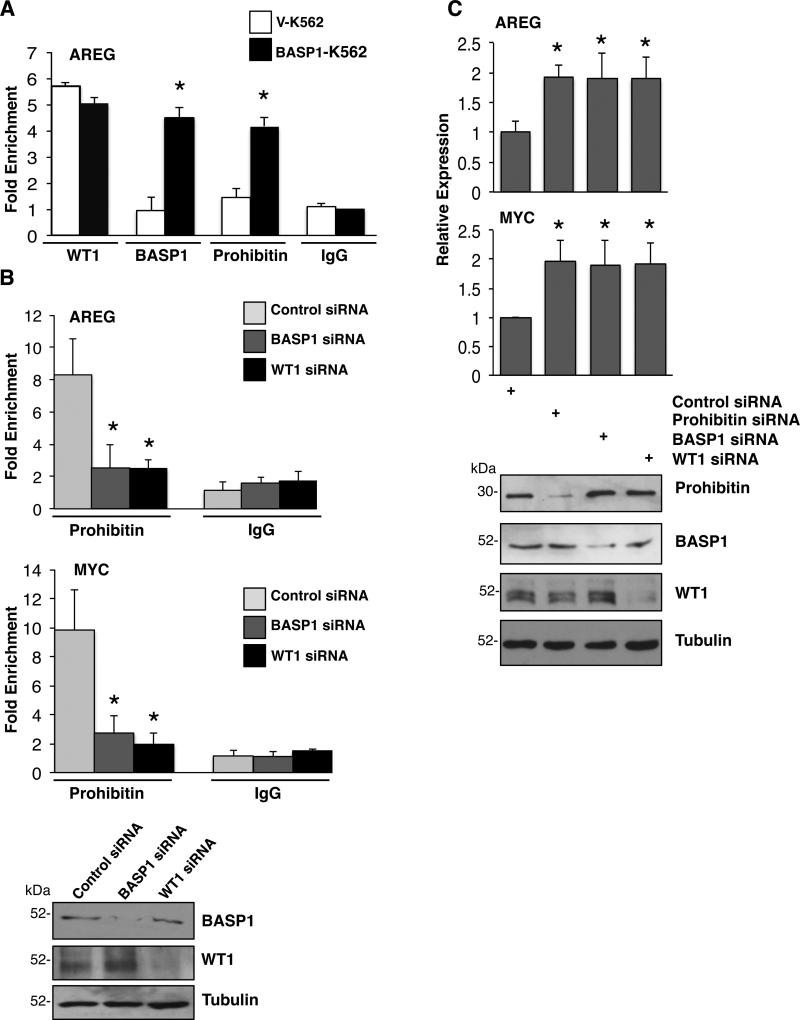
Figure 3. WT1-BASP1 dependent recruitment of Prohibitin to the promoters of WT1 target genes.
(A) Vector control K562 cells (V-K562) and BASP1-K562 cells were subjected to ChIP assay with control IgG, WT1, BASP1 or prohibitin antibodies. The data at the AREG promoter are presented as fold-enrichment relative to a control gene region. Error bars are obtained from standard deviation from the mean (SDM) of three independent experiments; *p<0.05 obtained by Student's t-test. (B) U2OS cells were transfected with control siRNA, BASP1 siRNA or WT1 siRNA and 48 hr later ChIP was performed with control IgG or anti-prohibitin antibodies. The data are presented as fold enrichment at the AREG and c-myc promoters relative to a control gene region. Error bars are obtained from SDM of three independent experiments; *p<0.05 obtained by Student's t test. Whole cell extracts were prepared parallel and immunoblotted with anti-prohibitin or anti-tubulin antibodies. (C) U2OS cells were transfected with a control siRNA, prohibitin siRNA, BASP1 siRNA or WT1 siRNA. 48 hr later RNA and cDNA were prepared and qPCR was performed to quantify GAPDH, AREG and c-myc mRNA. The data are presented relative to GAPDH mRNA. Values are the mean for three independent experiments with SDM and asterisk is student's t test of p<0.05. Whole-cell extracts were prepared in parallel and blotted with anti-prohibitin, anti-BASP1, anti-WT1 or anti-tubulin antibodies.