LETTER TO THE EDITOR
The ability to target myeloid malignancies using immunotherapy through means other than allogeneic transplantation depends on the capability to target leukemic clones while sparing normal tissues. It is now possible to generate clinical grade ex-vivo expanded T cells specific for leukemia associated antigens (LAA) for use in adoptive cell therapy(1). While a variety of putative LAAs in acute myeloid leukemia (AML) have been identified for use as potential targets for immunotherapy(2–8) and consensus panels have attempted to prioritize generic cancer antigens(9), a comprehensive evidence-based list of AML antigen targets has not yet been established. As a first step towards this we therefore analyzed, using quantitative real-time PCR (qRT-PCR), gene expression of 65 potential LAAs (Table S1) in de-identified, clinically annotated samples from 48 newly diagnosed untreated AML patients collected under IRB-approved protocols from three NCCN cancer centers.
A total of 52 samples (30 peripheral blood [PB], 22 bone marrow aspirate [BM]) from 48 AML patients were analyzed, including 4 patients for whom both PB and BM were available. The average age of the patients was 52 years (range 24–86); 52% were female. Seven patients had favorable cytogenetics while 11 were classified as adverse, 13 patients had FLT3 mutations (including 8 with FLT3-ITD), and 9 patients had mutations in NPM1 (Table S2; Table S3). RNA and DNA was isolated from the ficoll-purified PB and BM using AllPrep Mini Kits (Qiagen, Valencia, CA), and quantity, quality, and integrity of isolated RNA was assessed using a Nanodrop 1000 Spectrophotometer (Wilmington, DE) and Agilent RNA 6000 Nano Kit and 2100 Bioanalyzer (Santa Clara, CA). Only RNA with a RNA Integrity Number (RIN) of 7.0 or greater was used for subsequent analysis (Table S4). 400ng of high quality, total RNA was reverse-transcribed into cDNA using RT2 First Strand Kit (Qiagen). Custom RT2 Profiler PCR array plates (SABiosciences, Qiagen) were used for PCR reactions performed using RT2 SYBR Green ROX qPCR Mastermix (SABiosciences) according to the manufacturer’s instructions on an ABI 7900 thermal cycler (Applied Biosystems, Foster City, CA) with a program of 10 minutes at 95C, followed by 40 cycles at 95C for 15 seconds and 60C for 1 minute. Controls for human genomic DNA contamination, reverse transcription and PCR efficacy were included. Fold change expression values were calculated according to the comparative C(t) method(10). ΔC(t) was calculated as the C(t) of target gene “X” minus the geometric mean C(t) of reference genes HPRT1, PPIH, and TFRC in a sample. ΔC(t) for each target gene “X” in healthy donor samples were also computed in this manner. To calculate ΔΔC(t), median ΔC(t) of gene “X” in healthy donor blood or bone marrow (depending on the source of the AML sample) was subtracted from the ΔC(t) of X in the AML sample (ΔC(t) of X in AML sample – median ΔC(t) of X in healthy donor samples). Statistical analysis was performed using GraphPad Prism.
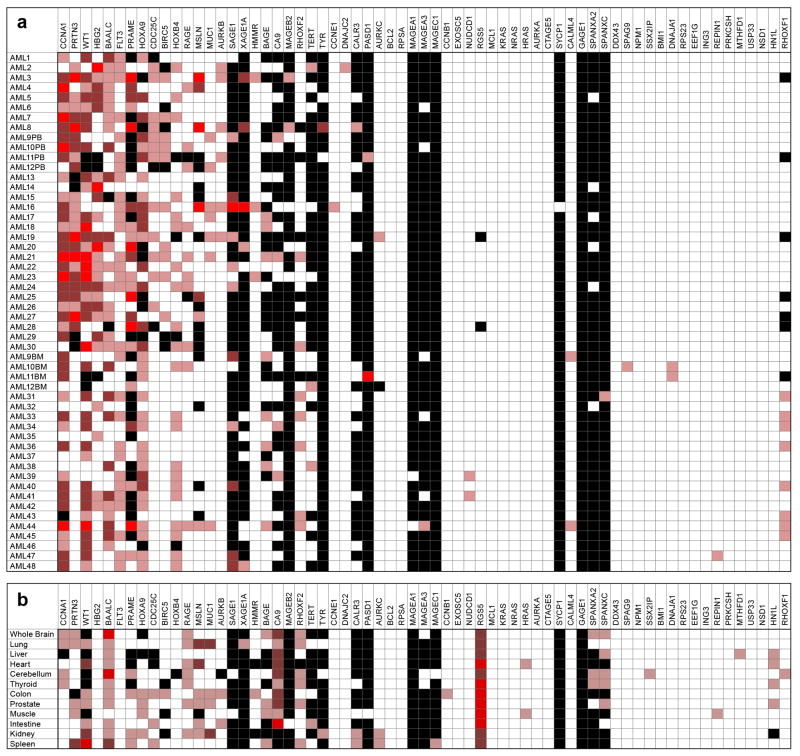
We observed considerable heterogeneity in levels of RNA overexpression of putative leukemia associated antigens compared to healthy donor tissues (Figure 1). Every AML sample had a least one LAA overexpressed, but no antigen was overexpressed in all AML samples. Surprisingly, the hemoglobin gamma globin gene HBG2, ordinarily expressed in the fetal liver, spleen and bone marrow but not usually in adulthood, which was recently identified as a leukemia antigen in a study of induced immune responses to GVAX/K562 vaccination in CML(11), was found to be frequently overexpressed in AML, often to a high level (Figure 1A; Figure 2). Similarly, CCNA1, WT1, BAALC, PR3 and PRAME were also highly overexpressed in multiple AML samples (Figure 2). We were able to confirm the previously reported (12)(13) association between FLT3-ITD mutated AML and overexpression of WT1 (Figure S2) but not other antigens. Finally, consistent with the fact that much of the existing evidence for myeloid LAA overexpression has been derived from the study of leukemic cell lines, we noted that the K562 human CML blast phase erythroleukemia cell line (ATCC, Manassas, VA) served as an excellent positive control for our panel with high expression of multiple previously described putative LAA including RHAMM, Survivin, h-TERT, CA9/CAIX, MAGEA3/6, MAGEB2, and MAGEC1 (Figure S1B) that were rarely detectable in our primary samples from AML patients (Figure 1A).
Figure 1. Expression of proposed leukemia associated antigens in acute myeloid leukemia (AML) patient samples and healthy tissues.
A) No single antigen was expressed in all cases of AML and many proposed antigen candidates are not frequently overexpressed in AML. PB=Peripheral Blood. BM=Bone Marrow. Fold change overexpression (OE) compared to median expression in healthy donors where light red indicates OE of 5–50x, red indicates OE of 50–500x, bright red indicates OE >500x. Black indicates no detectable expression; white indicates expression values seen in similar range as healthy donors. First thirty AML samples listed were from PB and are therefore compared to healthy donor PB, remaining 18 are from BM and compared to expression in healthy donor BM. B) Antigen expression in various human tissue types. Compared to median expression in healthy donors using same heat-map schema same as above.
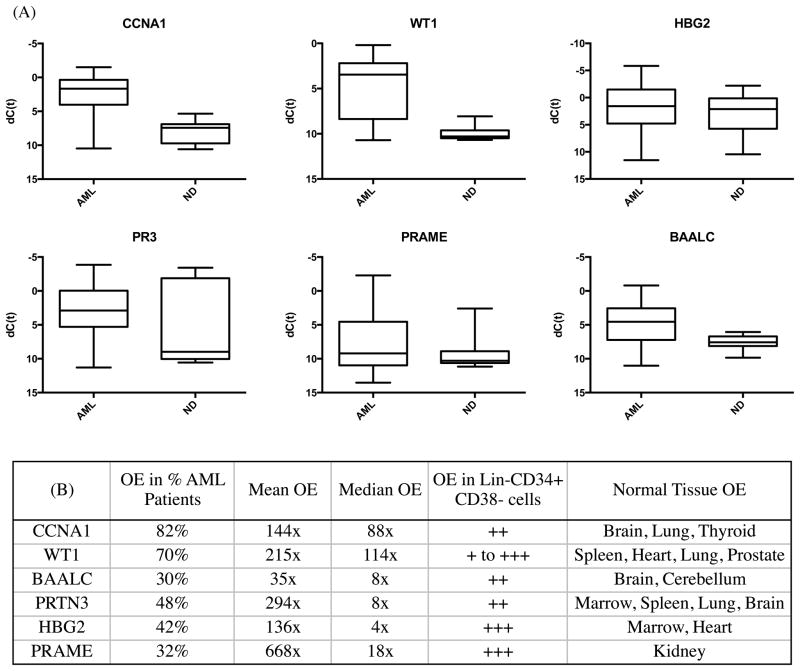
Figure 2. Top six acute myeloid leukemia associated antigens.
A) Therapeutic or diagnostic threshold for leading antigen candidates. Expression of antigens in all acute myeloid leukemia samples (AML) compared to all normal donor samples (ND) as box-plots where box represents 25th to 75th percentile and whiskers represent maximum and minimum dCT values observed. B) Detailed characteristics of leading AML antigen candidates including what percentage of AML samples tested had at least 5x overexpression (OE) of that gene, average levels of OE and OE in sorted lineage negative, CD34 positive and CD38 negative cells and normal donor tissues (see supplement for additional details).
The ideal targetable leukemia antigen would have high tumor-specific expression but no expression in healthy tissues(2). We therefore also quantified tissue expression of these putative LAAs in a range of normal tissues including samples (10 PB and 7 BM) from 17 healthy donors collected under IRB approved protocols at the NIH clinical center with an average age of 41 and 70% male, with ficoll purification and RNA extraction as described above. In addition, purified total RNA from a wide panel of human organs was obtained from Clontech (Mountain View, CA) (Table S5). Expression in at least some forms of healthy tissues was observed for almost all putative antigen targets (Figure 1B).
Several technical features are worthy of note. We found that samples with RNA integrity number (RIN) scores of less than 7.0 resulted in higher than expected C(t) outputs which correspond to lower gene expression when compared to samples with higher RIN scores (Figure S3) and were therefore excluded from analysis. Our array platform was highly sensitive and reproducible (Figure S1A), allowing for the reliable medium throughput analysis performed here. In most cases, antigen expression profiles from presentation blood and marrow samples from the same patient correlated closely (Figure S4). Gene expression of LAA in phenotypically identified AML blast populations sorted through flow cytometry did not markedly differ from the gene expression seen in the presentation peripheral blood sample from which they were isolated (Supplemental Methods; Table S6). Lastly, we were able to detect LAA overexpression across multiple samples from the same patient including unsorted peripheral blood samples, sorted AML blasts and a sorted (ie: Lineage negative, CD34 positive and CD38 negative) peripheral blood population enriched for stem cells (Supplemental Methods; Table S6).
This work has several obvious limitations. We performed this work on “real world” first presentation primary samples from three different leukemia centers in an attempt to limit biases introduced by presentation and referral patterns. Nevertheless, all institutions are highly specialized tertiary academic medical centers located in the northeastern and southeastern United States. While we do have age and gender patient demographics (Table S2), all these samples were from the first diagnosis before initiation of treatment for AML, and we unfortunately do not have any information on race or ethnic background, medical history including details on antecedent hematological conditions, prior/concurrent malignancies, or current medications with epigenetic or immune activity. We quantified total RNA expression levels (necessary but not sufficient for a targetable LAA) but did not provide information on protein expression or epitope processing and presentation by MHC; these factors will be addressed in future work now that the list of candidate AML LAAs has been substantially refined to exclude those not overexpressed in AML. Neo-antigens (ie: those generated by somatic mutations including single nucleotide variations, insertions, deletions and splice variants) are an important potential class of AML LAAs that were not investigated in this work, although extensive data on these AML specific sequence changes are now available and immune responses to epitopes created by these mutations have recently been described(14).
The ideal AML LAA would be expressed in most or all cases of AML but not in healthy tissues. Using a novel, highly sensitive and reproducible, real-time RT-PCR array testing only high quality RNA we show in this work that the majority of proposed “leukemia associated antigens” are expressed in the leukemia cell line K562 but often not in primary samples from AML patients. While we identified no antigen that was universally overexpressed in all AML samples, every patient did have at least one potentially targetable antigen overexpressed. We also noted significant healthy organ-specific tissue expression of many LAAs, highlighting the possibility of “off-target” effects, a finding not evident from the study of expression levels in peripheral blood and bone marrow alone. This list of genes overexpressed in AML, together with information regarding expression in a wide range of healthy tissue types, may be of use in AML as a reference for the selection of antigenic targets in adoptive T cell therapy and may also have utility in the PCR-based detection of minimal residual disease(15).
Supplementary Material
Acknowledgments
The authors thank Heidi Sardon, Ann Williams, Pradeep Dagur and J. Phillip McCoy of the Flow Cytometry Core Facility, National Heart, Lung, and Blood Institute, National Institutes of Health), Alan Hoofring of the NIH Medical Arts Service and the Johns Hopkins University Specimen Accessioning Core Lab for their assistance with this study.
This work was supported by the Intramural Research Program of the National Heart, Lung, Blood Institute of the National Institutes of Health.
Footnotes
Authorship: Contribution: MG and CSH designed and performed research, analyzed and interpreted data, and wrote the manuscript. TV and ML performed research. NH, BDS, GTP, SAS, MJ, BNS, JWF, HS, LQ, ATF, and HIL collected data, PM, SI, NAJ, MB, AJB interpreted data.
Supplementary information is available at Leukemia’s website.
Conflict-of-interest disclosure: HIL is an employee of Roche and a part-time faculty member at Johns Hopkins University. ATF serves on advisory boards for Seattle Genetics and Agios and has investigator initiated clinical trials currently funded by Exelexis, Seattle Genetics, and Millennium. All other authors report no relevant conflict of interest.
References
- 1.Weber G, Gerdemann U, Caruana I, Savoldo B, Hensel NF, Rabin KR, et al. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013. Mar 1, Generation of multi-leukemia antigen-specific T cells to enhance the graft-versus-leukemia effect after allogeneic stem cell transplant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anguille S, Van Tendeloo VF, Berneman ZN. Leukemia-associated antigens and their relevance to the immunotherapy of acute myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012 Oct;26(10):2186–2196. doi: 10.1038/leu.2012.145. [DOI] [PubMed] [Google Scholar]
- 3.el-Shami K, Smith BD. Immunotherapy for myeloid leukemias: current status and future directions. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008 Sep;22(9):1658–1664. doi: 10.1038/leu.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greiner J, Bullinger L, Guinn B-a, Doehner H, Schmitt M. Leukemia-Associated Antigens Are Critical for the Proliferation of Acute Myeloid Leukemia Cells. Clinical Cancer Research. 2008 Nov 15;14(22):7161–7166. doi: 10.1158/1078-0432.CCR-08-1102. [DOI] [PubMed] [Google Scholar]
- 5.Hourigan CS, Levitsky HI. Evaluation of current cancer immunotherapy: hemato-oncology. Cancer journal (Sudbury, Mass) 2011 Sep-Oct;17(5):309–324. doi: 10.1097/PPO.0b013e3182341fde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guinn BA, Gilkes AF, Woodward E, Westwood NB, Mufti GJ, Linch D, et al. Microarray analysis of tumour antigen expression in presentation acute myeloid leukaemia. Biochemical and biophysical research communications. 2005 Aug 5;333(3):703–713. doi: 10.1016/j.bbrc.2005.05.161. [DOI] [PubMed] [Google Scholar]
- 7.Atanackovic D, Luetkens T, Kloth B, Fuchs G, Cao Y, Hildebrandt Y, et al. Cancer-testis antigen expression and its epigenetic modulation in acute myeloid leukemia. American journal of hematology. 2011 Nov;86(11):918–922. doi: 10.1002/ajh.22141. [DOI] [PubMed] [Google Scholar]
- 8.Ochsenreither S, Majeti R, Schmitt T, Stirewalt D, Keilholz U, Loeb KR, et al. Cyclin-A1 represents a new immunogenic targetable antigen expressed in acute myeloid leukemia stem cells with characteristics of a cancer-testis antigen. Blood. 2012 Jun 7;119(23):5492–5501. doi: 10.1182/blood-2011-07-365890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009 Sep 1;15(17):5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 11.Qin L, Smith BD, Tsai HL, Yaghi NK, Neela PH, Moake M, et al. Induction of high-titer IgG antibodies against multiple leukemia-associated antigens in CML patients with clinical responses to K562/GVAX immunotherapy. Blood cancer journal. 2013;3:e145. doi: 10.1038/bcj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brackertz B, Conrad H, Daniel J, Kast B, Kronig H, Busch DH, et al. FLT3-regulated antigens as targets for leukemia-reactive cytotoxic T lymphocytes. Blood cancer journal. 2011 Mar;1(3):e11. doi: 10.1038/bcj.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greiner J, Schmitt M, Li L, Giannopoulos K, Bosch K, Schmitt A, et al. Expression of tumor-associated antigens in acute myeloid leukemia: Implications for specific immunotherapeutic approaches. Blood. 2006 Dec 15;108(13):4109–4117. doi: 10.1182/blood-2006-01-023127. [DOI] [PubMed] [Google Scholar]
- 14.Greiner J, Schneider V, Schmitt M, Götz M, Döhner K, Wiesneth M, et al. Immune responses against the mutated region of cytoplasmatic NPM1 might contribute to the favorable clinical outcome of AML patients with NPM1 mutations (NPM1mut) Blood 2013. 2013 Aug 8;122(6):1087–1088. doi: 10.1182/blood-2013-04-496844. [DOI] [PubMed] [Google Scholar]
- 15.Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nature reviews Clinical oncology. 2013 Aug;10(8):460–471. doi: 10.1038/nrclinonc.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.