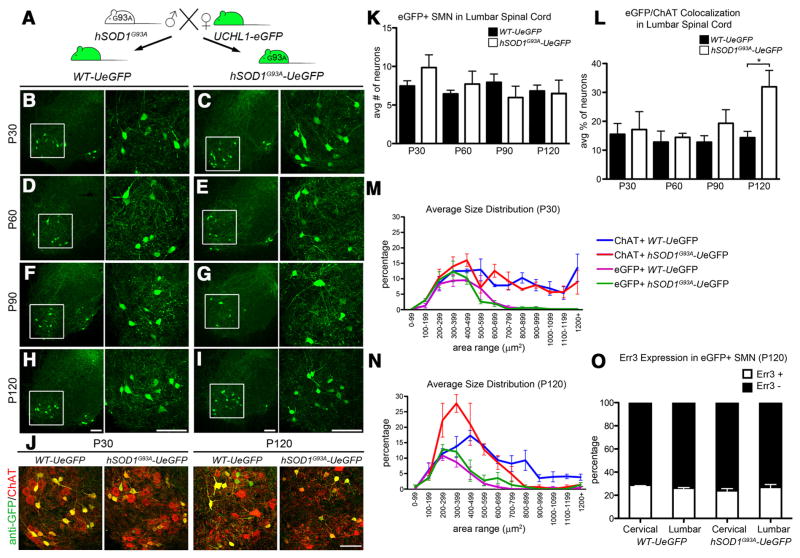
Figure 7.
eGFP+ neurons are more resistant to degeneration in the spinal cord of hSOD1G93A-UeGFP mice. A, hSOD1G93A transgenic males were mated with UCHL1-eGFP females to generate hSOD1pG93A-UeGFP mice and WT-UeGFP littermate controls. B–I, Representative images of lumbar ventral horns of age-matched WT-UeGFP (B,D,F,G) and hSOD1G93A-UeGFP (C,E,G,I) mice at P30 (B,C), P60 (D,E), P90 (F,G), and P120 (H,I). Boxed areas are enlarged to the right. J, Images of eGFP+ neurons in the ventral horn of lumbar spinal cord expressing ChAT at P30 and P120 in age-matched WT-UeGFP and hSOD1G93A-UeGFP mice. K, Bar graph representation of the average number of eGFP+ SMN in the lumbar spinal cord. L, Bar graph representation of the average eGFP+ /ChAT+ co-localization in lumbar spinal cord. M, N, Average size distribution of eGFP+ and ChAT+ SMN in cervical and lumbar spinal cord of age-matched WT-UeGFP and hSOD1G93A-UeGFP mice at P30 (M), and at P120 (N). O, Bar graph representation of percentage Err3 expression in eGFP+ SMN in age-matched WT-UeGFP and hSOD1G93A-UeGFP mice at P120. One-way ANOVA followed by Tukey’s post hoc multiple-comparison test and Student’s t test: *p < 0.05. Error bars indicate SEM. Scale bars: B–J, 100 μm.