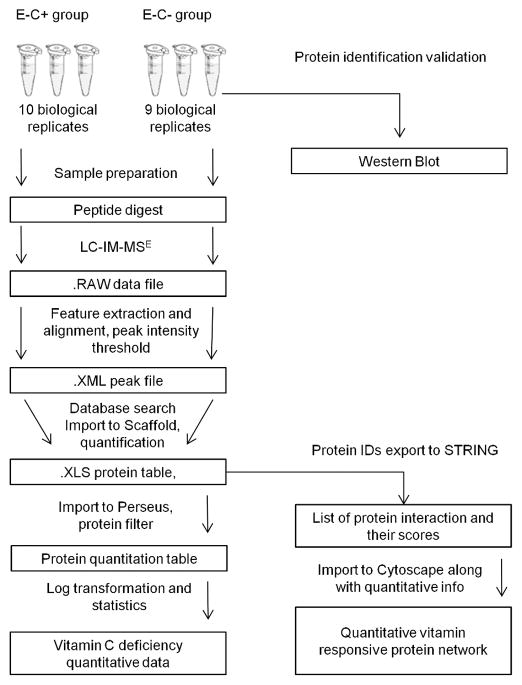
Figure 1.
Data analysis workflow for label-free quantitative proteomics using LC-IMS-MSE acquisitions. We implemented MassLynx and PLGS for peak extraction, alignment and database search, Scaffold for spectra validation and protein quantitation, Perseus for statistical analysis, and STRING and Cytoscape for protein interactions analysis and network construction.