Abstract
Cytochrome P450 enzymes are capable of catalyzing a great variety of synthetically useful reactions such as selective C-H functionalization. Surrogate redox partners are widely used for reconstitution of P450 activity based on the assumption that the choice of these auxiliary proteins or their mode of action does not affect the type and selectivity of reactions catalyzed by P450s. Herein, we present an exceptional example to challenge this postulate. MycG, a multifunctional biosynthetic P450 monooxygenase responsible for hydroxylation and epoxidation of 16-membered ring macrolide mycinamicins, is shown to catalyze the unnatural N-demethylation(s) of a range of mycinamicin substrates when partnered with the free Rhodococcus reductase domain RhFRED or the engineered Rhodococcus-spinach hybrid reductase RhFRED-Fdx. By contrast, MycG fused with the RhFRED or RhFRED-Fdx reductase domain mediates only physiological oxidations. This finding highlights the larger potential role of variant redox partner protein-protein interactions in modulating the catalytic activity of P450 enzymes.
Keywords: Cytochrome P450 enzymes, MycG, demethylation, redox partner, P450-reductase interaction
Introduction
The superfamily of cytochrome P450 enzymes (CYPs) is one of the most versatile biocatalyst systems in nature1-3. CYPs are capable of catalyzing more than twenty distinct types of reactions including regio- and stereoselective oxidation of unactivated C-H bonds (hydroxylation and epoxidation), dealkylation, C-C bond cleavage, aromatic coupling and others4,5. Recently, more novel activities such as decarboxylation6, nitration7, farnesene synthase activity8, and carbene transfer9 have been reported, reflecting the ability to further diversify the function of these versatile enzymes.
Catalytic activities of the vast majority of CYPs require one or more redox partner proteins to sequentially deliver two electrons from NAD(P)H to the heme iron reactive center for dioxygen activation1,2. Classically, there are two major redox partner systems10,11, including (1) a two-component system for most bacterial and mitochondrial CYPs comprising a small iron-sulfur redoxin and an FAD-containing redoxin reductase, and (2) a single FAD/FMN-containing CYP reductase that serves in eukaryotic microsomal P450s as a separate partner protein, or in a small number of bacterial CYPs (e.g. P450BM3) as a fused functional domain. Interestingly, non-classical redox systems have been discovered continuously10,12, among which the FMN/Fe2S2 containing reductase domain termed “RhFRED” that is naturally fused with the P450 enzyme from Rhodococcus sp. NCIMB 978413 has been extensively studied in our laboratories14-16.
For practical purposes due to the difficulty of obtaining native partners, one or more surrogate redox partners, acting either in isolation or as artificially fused protein complexes, are often employed in functional characterization or synthetic applications of CYPs. The choice of surrogate partners or their mode of action is not necessarily expected to affect the type and selectivity of reactions catalyzed by P450s. Although alternative redox partners may influence catalytic efficiency and/or product distribution17-22, the chemical identity of products themselves is apparently determined by the P450 enzyme. Our previous experience constructing and characterizing various self-sufficient biosynthetic P450 enzymes (whose activity is independent of isolated redox partners) fused with the RhFRED reductase domain14-16 also supports this “postulate”.
Notably, previous work revealed that cytochrome b5 is able to modulate the bi-functional role of human CYP17A1 as a 17α-hydroxylase or a 17,20-lyase in steroid biosynthesis23. This well-studied example demonstrates that the catalytic type of a P450 enzyme can be altered by a third-party protein effector via alternative interactions among the P450 enzyme, P450 reductase, effector protein, and distinct substrates24-27. However, there have been no reports for a redoxin protein or a P450 reductase acting as the specific effector by itself to endow the serving P450 enzyme with new catalytic activity.
In this work, novel demethylated mycinamicin products were generated by P450 MycG supported by a stand-alone form of the RhFRED reductase domain, as opposed to RhFRED fused to MycG, thus challenging the generally accepted postulate. Further engineering of RhFRED led to didemethylated mycinamicin macrolides, demonstrating a novel route for structural diversification of natural products. Mechanistically, our results suggest that variant protein-protein interactions with the redox partner may serve as a modulator of P450 function by dramatically affecting the substrate binding mode in the P450 active site.
Results
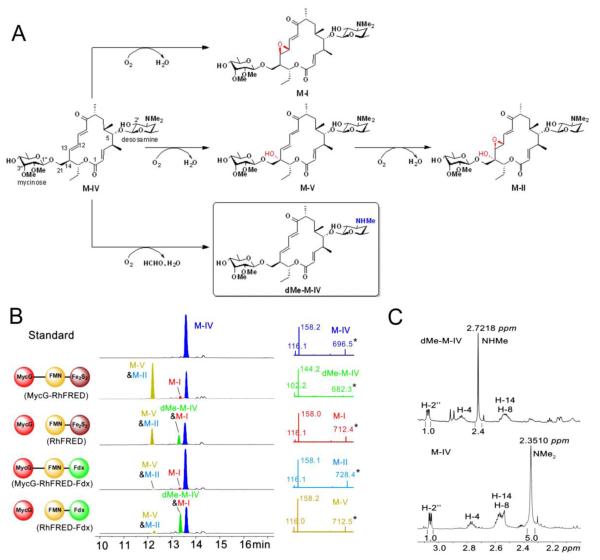
MycG is the multifunctional P450 monooxygenase involved in the biosynthetic pathway of 16-membered ring macrolide mycinamicins in the rare actinomycete Micromonospora gris-eorubida19,28,29. Physiologically, it catalyzes the C14 hydroxylation, and the C12=C13 epoxidation of mycinamicin IV (MIV) leading to mycinamicin V (M-V) and mycinamicin I (M-I), respectively. M-V is subsequently epoxidized by MycG, giving rise to the major final product mycinamicin II (M-II), while M-I cannot be further modified by MycG, thus representing the other terminal product in the pathway (Figure 1A). All diglycosylated mycinamicins display strong antibiotic activity against Gram-positive bacteria and mycoplasma30, and M-II has been developed into a veterinary anti-infective agent31.
Figure 1.
Oxidation and demethylation of M-IV catalyzed by MycG using different redox systems (2 h reactions). (A) Scheme for MycG catalyzed reactions. The novel demethylation product dMe-M-IV is shown in box. The introduced hydroxyl and epoxy groups are labelled in red. The demethylated group is highlighted in blue. (B) Left panel: LC traces at 280 nm of different reaction extracts. The redox systems used are shown at the left side of the corresponding traces. Mycinamicin derivatives are colored differently for clarity. Due to close polarity, M-V and M-II were co-eluted; dMe-M-IV and M-I were co-eluted. Since M-I and M-II lacking of the diene moiety only have weak absorbance at 280 nm, the formation of products appear not to be proportional to substrate consumption. Right panel: MS/MS analysis of each mycinamicin compound (see Figure S1 for explanations of the secondary mass spectra). (C) Comparison of the 1H NMR spectra (see Figure S2 and S3 for full spectra) of M-IV and dMe-M-IV. The chemical shift and integrated peak area of the N-monomethyl group in dMe-M-IV are apparently different from those of the N-dimethyl group in M-IV.
The multifunctional activities of MycG were previously reconstituted in vitro by using the commercial surrogate redox partners spinach ferredoxin (Fdx) and spinach ferredoxin-NADP+ reductase (FdR), since the native redox system driving catalytic activity of MycG remains unknown19. Motivated by our recent work14-16 and that of others32-35 in enhancing catalytic activity by fusing the RhFRED reductase domain to the C-terminus of various biosynthetic P450s, we constructed the MycG-RhFRED fusion protein. Similar to MycG supported by separate spinach Fdx/FdR, this single-component self-sufficient P450 system driven by NADPH, achieved all physiological reactions known for MycG (Figure 1B).
To investigate if the free form of the RhFRED domain can support the catalytic function of MycG, the ability of this two-component system to convert M-IV was tested. Surprisingly, in addition to the expected oxidative products, M-I, M-II, and M-V (co-eluted with M-II), a new product formed with 18.5% isolated yield (Table 1) with the same retention time as M-I was observed in LC-MS analysis of the reaction mixture (Figure 1B). Because of its strong absorbance at 280 nm, resembling that of M-IV and M-V, but distinct from M-I, we assumed that the chemical structure of the new compound likely retained an intact diene moiety. Indeed, its m/z value of 682.3 is 14 amu less than that of protonated M-IV, suggesting a demethylated product, which was also supported by high resolution mass spectrometry ([M+H]+: obs. 682.4161, calc. 682.4160). Furthermore, the MS/MS spectrum of the demethylated M-IV (dMe-MIV) showed two fragmentation ions of 144.2 and 102.2, consistent with a demethylated desosamine moiety, while the corresponding fragmentation ions for intact desosamine are 158.2 and 116.115,36 (Figure 1B and S1). Detection of formaldehyde in the reaction by Purpald reagent37 provides additional evidence for oxidative demethylation (see Supporting Information). To confirm the site of demethylation, the 1H NMR of enzymatically prepared dMe-MIV was analyzed using M-IV as reference (Figure 1C, S2 and S3). As expected, the 2.35 ppm singlet peak of M-IV that corresponds to the N-dimethyl group disappeared in the spectrum of dMe-MIV. Instead, a new singlet peak was observed with half the integrated area at 2.72 ppm, indicating the —NHMe group resulted from the MycG-catalyzed demethylation.
Table 1.
Quantitative measurement of M-IV conversions mediated by different MycG catalytic systems (2 h reactions)
| Relative AUC280nma of oxidized products |
Relative AUC280nm of deMe-IV |
Isolated yield of deMe-IV |
Rate constant (k, min−1) |
Coupling efficiencyb | |
|---|---|---|---|---|---|
| MycG + RhFRED | 22.1 ± 1.1% | 27.9 ± 1.4% | 18.5% | 0.036 ± 0.008 | 29.4 ± 0.8% |
| MycG + RhFRED-Fdx | 5.8 ± 0.8% | 34.5 ± 1.5% | 24.0% | 0.036 ± 0.007 | 24.2 ± 0.9% |
| MycG-RhFRED | 54.1 ± 2.0% | - | - | 0.052 ± 0.012 | 18.7 ± 0.5% |
| MycG-RhFRED-Fdx | 5.0 ± 0.4% | - | - | 0.017 ± 0.004 | 21.3 ± 0.7% |
The numbers of relative AUC280nm (areas under curve at 280 nm) were calculated from the integrated area of certain peaks on HPLC traces
Coupling efficiencies were calculated as the percentage of NADPH used for product formation over the total NADPH consumption
This observation is intriguing because a heretofore unknown dMe-MIV product was obtained following separation of the previously fused protein domains, MycG and RhFRED. Next, we measured the oxidation and demethylation activities of MycG under a variety of molar ratios (1:1, 1:10, and 1:100) between MycG and RhFRED. In all cases, both oxidative and demethylated products were detected (Figure S4), indicating the ability of MycG to catalyze demethylation is not the consequence of a stoichiometric effect of RhFRED. A time course of the reaction (Figure S5) showed proportional accumulation of oxidative and demethylated mycinamicin products.
Sequence analysis shows that RhFRED consists of two functional domains, the FMN domain and the Fe2S2 domain38,39. To analyze the impact of each domain on demethylation by MycG, the Fe2S2 domain of RhFRED, which is presumably involved in direct interaction with the P450 enzyme, was exchanged for the spinach Fdx, yielding a hybrid Rhodococcus-spinach reductase, RhFRED-Fdx. When partnered with this hybrid reductase, the MycG-catalyzed reaction produced even greater amounts of dMe-MIV (24.0% isolated yield, see Table 1), while the oxidized products M-I, M-V, and M-II were also detectable (Figure 1B). This is in contrast to the MycG-RhFRED-Fdx fusion system which, similarly to MycG-RhFRED, generated only oxidized products, albeit in lower quantities. These results suggest that MycG-mediated demethylation is mainly determined by the mode of interaction between MycG and its redox partner rather than by the redox partner itself.
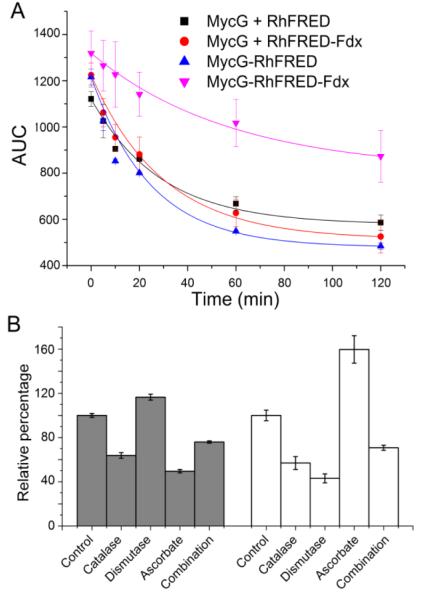
Quantitatively, by fitting the substrate consumption data into the one phase exponential decay curve, the rate constants (k) of the four studied MycG catalytic systems were determined (Table 1 and Figure 2A). In comparison, isolated RhFRED and RhFRED-Fdx served MycG with close efficiency despite of distinct product profiles. The M-IV oxidation mediated by MycG-RhFRED and MycG-RhFRED-Fdx demonstrated the apparent reaction velocity at 500 μM of M-IV being 26.0 ± 6.0 μM min−1 and 8.5 ± 2.0 μM min−1 (k × [S]), respectively. The steady-state Michaelis-Menten kinetics of MycG could not be ascertained due to multiple reactions occurring simultaneously.
Figure 2.
(A) Enzymatic consumption of M-IV (calculated from HPLC areas under curve, AUC) fit into the one phase exponential decay curve. The rate constants (k) are shown in Table 1. (B) The effects of catalase (20 U), superoxide dismutase (2 U), ascorbate (10 mM), and the combination of these three ROS scavengers on the activity of MycG/RhFRED against M-IV. Closed bar, the ratio of demethylated product/oxidized products relative to that of the control reaction without addition of any scavengers; open bar, the overall conversion percentage calculated based on the substrate M-IV consumption. The percentage numbers from the control reaction are arbitrarily assigned to be 100%.
It is well known that the use of an unnatural redox partner in a CYP catalyzed reaction likely uncouples the electron generation upon NAD(P)H consumption and P450 product formation40-42. Separation of the fused P450 and reductase domains could have a similar effect43. The uncoupling process generates reactive oxygen species (ROS) such as superoxide anions (O −2) and H2O2, which could direct the P450 catalytic route into the “peroxide shunt pathway”1,2. To examine wheth er the demethylation of mycinamicins is a consequence of the peroxygenase activity by undergoing this shunt pathway, we first determined the NADPH coupling efficiency of the four catalytic systems using M-IV as substrate. As shown in Table 1, the coupling efficiencies for P450-reductase fusions (18.7% for MycG-RhFRED and 21.3% for MycG-RhFRED-Fdx) were lower than those of corresponding reactions using separated reductases (29.4% for MycG + RhFRED and 24.2% for MycG + RhFRED-Fdx). These data indicate that separation of reductase and P450 domains did not result in greater uncoupling in these P450 catalytic systems.
Moreover, catalase, superoxide dismutase (SOD), ascorbic acid, and the combination of these three ROS scavengers were added into the MycG/RhFRED/M-IV reaction to remove ROS possibly generated from the uncoupling process. Although these ROS scavengers lowered the M-IV conversion (except for ascorbate), as well as the ratio of demethylated product to oxidized products (except for SOD) in the majority of reactions (Figure 2B), these effects were not dose-dependent, and the N-demethylation activity remained even at the highest concentration of catalase, SOD, and ascorbate (Figure S6).
These results, together with the small change of coupling efficiency upon reductase domain separation, suggest that dMe-M-IV is unlikely to be a product of the peroxide shunt pathway. However, the lowered demethylated product/oxidized product ratio implies superoxide species might still contribute to some of the observed demethylation. This could be supported by detection of a low level of H2O2 in MycG reactions (Figure S7), and the fact that addition of ascorbic acid improved the overall conversion of M-IV (Figure 2B) since this chemical scavenger is capable of protecting the enzyme and substrate/products from radical damage by superoxides44.
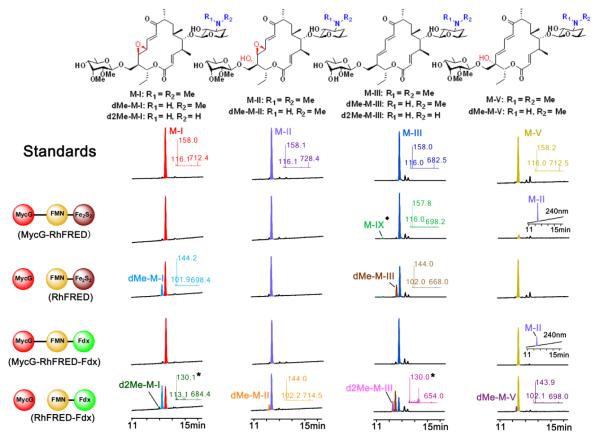
Next, we comparatively tested the activity of MycG, driven by each of the four different redox systems, against M-IV analogues M-I, II, III, and V, isolated from fermentation cultures of wild-type or mutant M. griseorubida30,45. The two fused systems displayed the typical physiological set of the hydroxylation and/or epoxidation products (Figure 3). However, the reactions driven by free redox partners gave rise to more diversity of demethylated products. Specifically, both RhFRED and RhFRED-Fdx efficiently supported the demethylation of M-I and M-III by MycG, generating the new products dMe-MI and dMe-M-III, respectively. M-II and M-V were demethylated by MycG at a low level driven by RhFRED-Fdx, but not by RhFRED. Of particular interest, MycG partnered by the stand-alone form of RhFRED-Fdx was able to further remove the second N-methyl group from mono-demethylated dMe-MI and dMe-M-III, leading to the unique double-demethylated product d2Me-M-I and d2Me-M-III (Figure 3), respectively. Previously, both in vivo30,45 and in the in vitro reactions that employed the surrogate spinach Fdx/FdR system19, M-I and M-II have been shown to be the two end products of the mycinamicin biosynthetic pathway, and M-III bearing the monomethoxy sugar javose is oxidized by MycG at a very low level. Here, the change of the redox partners and their interaction mode has led to seven novel demethylated products (Figure 1, 3, and S8), demonstrating a new route for structural diversification of natural products.
Figure 3.
LC-MS analysis of the MycG reactions using M-I, M-II, M-III, and M-V as substrates. The structures of substrates and products are shown on top. The reactions catalyzed by the same enzyme(s) are aligned in each row. The reactions using the common substrate are arranged in the same column. The LC traces of M-I and M-II were recorded at 240 nm. The LC traces of M-III and M-V were recorded at 280 nm. In the M-V reactions catalyzed by two fusion MycG enzymes, the accumulation of M-II (seen at 240 nm) that is co-eluted with the unreacted M-V is shown in insets. The selected MS/MS results are displayed in insets, whose colors are consistent with those of the corresponding LC peaks. The asterisked numbers indicate the mass for the secondary ion fragment of double-demethylated mycinamicins (Figure S1). The structure of M-IX (the hydroxylated M-III labelled by the diamond symbol) is shown in Figure S8.
Discussion
In this study we show for the first time the ability of the multifunctional P450 monooxygenase MycG to catalyze N-demethylation of various mycinamicin natural product molecules. This novel functionality of MycG occurs when the monooxygenase is partnered with either the free Rhodococcus reductase domain, RhFRED, or the Rhodococcus-spinach hybrid, RhFRED-Fdx. In contrast, no similar demethylation activity was observed with the corresponding fused redox partner systems. The minor differences in the NADPH coupling efficiency upon P450-reductase domain separation (Table 1), and dose-independent effects of catalase, SOD, and the chemical radical scavenger ascorbate on both MycG activity and the demethylation/oxidation product distribution (Figure 2B), strongly suggest that the unnatural demethylation activity is primarily due to heme-iron-oxo catalysis, as opposed to the peroxide shunt pathway. Thus, this finding highlights a broader role of redox partner protein-protein interactions in modulating catalytic activity of the P450 enzyme. It is well known that alternative or surrogate redox partner protein(s) might not provide the optimal supporting activity toward mismatched P450 enzymes17,19,22, and could influence product distribution14,16. The modulating effect of a third party protein, cytochrome b5, on the hydroxylase/lyase bi-function of CYP17A1 supported by cytochrome P450 reductase23-26 has also been demonstrated. However, to the best of our knowledge, the entirely new catalytic activity of a P450 enzyme associated with an alternative redox partner is unprecedented.
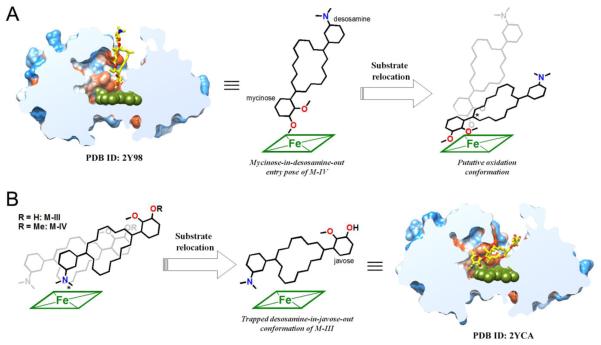
The novel catalytic activity demonstrated by MycG likely stems from the poor accessibility of the substrate binding site, as evidenced by the x-ray structure analysis of the MycG-substrate complexes46. Thus, the bulky and conformationally restrained mycinamicin substrates were largely unable to penetrate the L-shaped binding site to adopt a catalytically productive binding mode. NMR modelling using paramagnetic restraints suggests displacement of a number of the protein secondary structure elements takes place in order to accommodate mycinamicin substrates in an orientation conducive to the observed oxidation pattern46. By analogy to P450cam47,48 and the human P450 3A449, reorganization of the MycG active site may be driven by binding of the redox partner. As has been recently demonstrated by elegant work from the Poulos laboratory35, binding of the endogenous redox partner putidaredoxin stabilizes the “open” conformation of P450cam, facilitating substrate access to the active site.
Consistent with the structurally resolved MycG-substrate complexes46, mycinamicins exhibiting pseudo two-fold symmetry enter the active site in one of the two possible orientations: “mycinose-in-desosamine-out” (Figure 4A) or “desosa-mine-in-mycinose-out” (Figure 4B). The desosamine-in-mycinose-out entry results in the catalytically non-productive binding mode represented by the M-III co-structure (PDB ID code 2YCA). Remarkably, the mycinose-in-desosamine-out entry in the absence of the redox partner resulted in both the M-IV and M-V substrates being stalled in the intermediate non-productive binding pose in all the structurally resolved complexes46. If substrate relocation to the catalytically competent mode is indeed initiated by specific interactions with the endogenous redox partner, the alternative surrogates may to some extent mimic this allosteric effect, either facilitating or hampering substrate progression to the catalytic site. Tethered to MycG, both RhFRED and RhFRED-Fdx afforded physiological product profiles, with the qualification that the product yield was low for MycG-RhFRED-Fdx. Alternatively, when used as stand-alone redox partners, both the RhFRED and RhFRED-Fdx domains produced demethylated (RhFRED) or di-demethylated (RhFRED-Fdx) products of not only the native MycG substrates M-IV and M-V, but also the M-III precursor, an otherwise poor substrate for MycG oxidation, which was efficiently demethylated (Figure 3).
Figure 4.
Dual binding modes of the mycinamicin substrates in the active site of MycG. (A) A putative binding pose of M-IV leading to physiological C14 hydroxylation or/and C12-C13 epoxidation, which might be derived from the non-catalytic “mycinose-in-desosamineout” entry pose as observed in the 2Y98 (PDB ID) structure. (B) A putative “desosamine-in-mycinose/javose-out” binding pose yielding N-demethylation is likely to be induced or stabilized by protein-protein interactions between MycG and separate redox partners. The non-catalytic conformation observed in the 2YCA structure may be derived from this pose. The mycinamicin chemical structures are simplified for clarity. Oxygen atoms are shown in red and nitrogen in blue. Heme is shown in dark green. The reactive sites are asterisked. Slice through the MycG binding site shows M-IV or M-III in virtually orthogonal orientations experimentally observed in the crystal structures46. Protein surface is colored by hydrophobicity, hydrophobic areas are in orange and hydrophilic areas in blue.
To explain this phenomenon we speculate that while the electron transfer between MycG and the surrogate redox partners is retained, the allosteric effect facilitating substrate progression to the normal catalytically competent binding mode (“mycinose-in-desosamine-out”) is likely diminished, if not entirely lost. This might be due to intrinsically low affinity between the surrogate partners and MycG, particularly if concentration dependence in binary complex formation becomes a rate-limiting issue in the bimolecular reaction. Also, loss of the positioning effect of the linker following domain separation may be a factor in changing the interaction mode of free reductase and P450 domains. We conclude that in the absence of the allosteric influence of the redox partner, mycinamicins are more inclined to be stalled in the “desosamine-in-mycinose-out” non-catalytic entry pose, increasing the probability of N-demethylation occurring in an otherwise atypical reaction site.
Despite discovery of a new N-demethylation activity for monooxygenase MycG, caution should be exercised in interpreting reconstituted P450 activity generated in a surrogate redox system, particularly until the generality of this approach is further investigated. However, an interesting hypothesis is suggested: P450 enzymes could be even more versatile in vivo than previously considered, because these biocatalysts might interact with a variety of redox partners to gain alternative activities. This may impart evolutionary advantages to the host organisms through detoxifying a more diverse range of xenobiotics or synthesizing more secondary metabolites to adapt to ever-changing environments. Clear indications that the P450-omes of microbes (e.g. Streptomyces coelicolor17,22) are capable of interacting with a limited set of redox partner are consistent with a broader role for redoxin-mediated selectivity in these fascinating enzyme systems.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by funding from “Recruitment Program of Global Experts, 2012” (S.L.) and NIH grant GM078553 (D.H.S. and L.M.P.). We are also grateful for the supports from National Natural Science Foundation of China (NSFC 31270855 to S.L. and NSFC 31300075 to W.Z.). We thank Mr. Potter Wickware for his attentive reading of the manuscript.
ABBREVIATIONS
- M-I-V
mycinamicin I-V
- M-IX
mycinamicin IX
- dMe-M-I-V
N-demethylated mycinamicin I-V
- d2Me-M-I
N-didemethylated mycinamicin I
- d2Me-M-III
N-didemethylated mycinamicin III
Footnotes
Supporting Information Experimental methods, DNA sequences for the synthesized fdx gene and the gene encoding the RhFRED-Fdx hybrid protein, NMR spectra, the data for effects of catalase, superoxide dismutase, ascorbate on MycG activity, and etc. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd Ed Kluwer Academic/Plenum Publishers; New York: 2005. [Google Scholar]
- (2).Coon MJ. Annu. Rev. Pharmacol. Toxicol. 2005;45:1. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- (3).Podust LM, Sherman DH. Nat. Prod. Rep. 2012;29:1251. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Guengerich FP. Chem. Res. Toxicol. 2001;14:611. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- (5).Guengerich FP, Munro AW. J. Biol. Chem. 2013;288:17065. doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rude MA, Baron TS, Brubaker S, Alibhai M, del Cardayre SB, Schirmer A. Appl. Environ. Microbiol. 2011;77:1718. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Barry SM, Kers JA, Johnson EG, Song L, Aston PR, Patel B, Krasnoff SB, Crane BR, Gibson DM, Loria R, Challis GL. Nat. Chem. Biol. 2012;8:814. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR. J. Biol. Chem. 2009;284:36711. doi: 10.1074/jbc.M109.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Coelho PS, Brustad EM, Kannan A, Arnold FH. Science. 2013;339:307. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- (10).Munro AW, Girvan HM, McLean KJ. Nat. Prod. Rep. 2007;24:585. doi: 10.1039/b604190f. [DOI] [PubMed] [Google Scholar]
- (11).Hannemann F, Bichet A, Ewen KM, Bernhardt R. Biochim. Biophys. Acta. 2007;1770:330. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- (12).Munro AW, Girvan HM, McLean KJ. Biochim. Biophys. Acta. 2007;1770:345. doi: 10.1016/j.bbagen.2006.08.018. [DOI] [PubMed] [Google Scholar]
- (13).Roberts GA, Grogan G, Greter A, Flitsch SL, Turner NJ. J. Bacteriol. 2002;184:3898. doi: 10.1128/JB.184.14.3898-3908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Li S, Podust LM, Sherman DH. J. Am. Chem. Soc. 2007;129:12940. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li S, Chaulagain MR, Knauff AR, Podust LM, Montgomery J, Sherman DH. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18463. doi: 10.1073/pnas.0907203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Carlson JC, Li S, Gunatilleke SS, Anzai Y, Burr DA, Podust LM, Sherman DH. Nat. Chem. 2011;3:628. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chun YJ, Shimada T, Sanchez-Ponce R, Martin MV, Lei L, Zhao B, Kelly SL, Waterman MR, Lamb DC, Guengerich FP. J. Biol. Chem. 2007;282:17486. doi: 10.1074/jbc.M700863200. [DOI] [PubMed] [Google Scholar]
- (18).Sadeghi SJ, Meharenna YT, Fantuzzi A, Valetti F, Gilardi G. Faraday Discuss. 2000;116:135. doi: 10.1039/b003180l. [DOI] [PubMed] [Google Scholar]
- (19).Anzai Y, Li S, Chaulagain MR, Kinoshita K, Kato F, Montgomery J, Sherman DH. Chem. Biol. 2008;15:950. doi: 10.1016/j.chembiol.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. J. Biol. Chem. 2006;281:26289. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Harada H, Shindo K, Iki K, Teraoka A, Okamoto S, Yu F, Hattan J.-i., Utsumi R, Misawa N. Appl. Microbiol. Biotechnol. 2011;90:467. doi: 10.1007/s00253-010-3062-9. [DOI] [PubMed] [Google Scholar]
- (22).Lei L, Waterman MR, Fulco AJ, Kelly SL, Lamb DC. Proc. Natl. Acad. Sci. U. S. A. 2004;101:494. doi: 10.1073/pnas.2435922100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lee-Robichaud P, Wright JN, Akhtar ME, Akhtar M. Biochem. J. 1995;308:901. doi: 10.1042/bj3080901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Akhtar MK, Kelly SL, Kaderbhai MA. J. Endocrinol. 2005;187:267. doi: 10.1677/joe.1.06375. [DOI] [PubMed] [Google Scholar]
- (25).DeVore NM, Scott EE. Nature. 2012;482:116. doi: 10.1038/nature10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Estrada DF, Laurence JS, Scott EE. J. Biol. Chem. 2013;288:17008. doi: 10.1074/jbc.M113.468926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Bridges A, Gruenke L, Chang Y-T, Vakser IA, Loew G, Waskell L. J. Biol. Chem. 1998;273:17036. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- (28).Anzai Y, Saito N, Tanaka M, Kinoshita K, Koyama Y, Kato F. FEMS Microbiol. Lett. 2003;218:135. doi: 10.1111/j.1574-6968.2003.tb11509.x. [DOI] [PubMed] [Google Scholar]
- (29).Inouye M, Takada Y, Muto N, Beppu T, Horinouchi S. Mol. Gen. Genet. 1994;245:456. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- (30).Satoi S, Muto N, Hayashi M, Fujii T, Otani M. J. Antibiot. 1980;33:364. doi: 10.7164/antibiotics.33.364. [DOI] [PubMed] [Google Scholar]
- (31).Takenaka S, Yoshida K, Yamaguchi O, Shimizu K, Morohoshi T, Kinoshita K. FEMS Microbiol. Lett. 1998;167:95. doi: 10.1111/j.1574-6968.1998.tb13213.x. [DOI] [PubMed] [Google Scholar]
- (32).Robin A, Roberts GA, Kisch J, Sabbadin F, Grogan G, Bruce N, Turner NJ, Flitsch SL. Chem. Comm. 2009;2009:2478. doi: 10.1039/b901716j. [DOI] [PubMed] [Google Scholar]
- (33).Sabbadin F, Hyde R, Robin A, Hilgarth EM, Delenne M, Flitsch SL, Turner NJ, Grogan G, Bruce NC. Chembiochem. 2010;11:987. doi: 10.1002/cbic.201000104. [DOI] [PubMed] [Google Scholar]
- (34).Fujita N, Sumisa F, Shindo K, Kabumoto H, Arisawa A, Ikenaga H, Misawa N. Biosci. Biotechnol. Biochem. 2009;73:1825. doi: 10.1271/bbb.90199. [DOI] [PubMed] [Google Scholar]
- (35).Robin A, Kohler V, Jones A, Ali A, Kelly PP, O’Reilly E, Turner NJ, Flitsch SL. Beilstein J. Org. Chem. 2011;7:1494. doi: 10.3762/bjoc.7.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Harada K-I, Takeda N, Suzuki M, Hayashi M, Ohno M, Satoi S. J. Antibiot. 1985;38:868. doi: 10.7164/antibiotics.38.868. [DOI] [PubMed] [Google Scholar]
- (37).Zhang K, Damaty SE, Fasan R. J. Am. Chem. Soc. 2011;133:3242. doi: 10.1021/ja109590h. [DOI] [PubMed] [Google Scholar]
- (38).Roberts GA, Celik A, Hunter DJ, Ost TW, White JH, Chapman SK, Turner NJ, Flitsch SL. J. Biol. Chem. 2003;278:48914. doi: 10.1074/jbc.M309630200. [DOI] [PubMed] [Google Scholar]
- (39).Hunter DJ, Roberts GA, Ost TW, White JH, Müller S, Turner NJ, Flitsch SL, Chapman SK. FEBS Lett. 2005;579:2215. doi: 10.1016/j.febslet.2005.03.016. [DOI] [PubMed] [Google Scholar]
- (40).Ba L, Li P, Zhang H, Duan Y, Lin Z. Biotechnol. Bioeng. 2013;110:2815. doi: 10.1002/bit.24960. [DOI] [PubMed] [Google Scholar]
- (41).Harskamp J, Britz-McKibbin P, Wilson JY. Anal. Chem. 2012;84:862. doi: 10.1021/ac202787n. [DOI] [PubMed] [Google Scholar]
- (42).Jensen K, Johnston JB, Ortiz de Montellano PR, Møller BL. Biotechnol. Lett. 2012;34:239. doi: 10.1007/s10529-011-0768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Girvan HM, Waltham TN, Neeli R, Collins HF, McLean KJ, Scrutton NS, Leys D, Munro AW. Biochem. Soc. Trans. 2006;34:1173. doi: 10.1042/BST0341173. [DOI] [PubMed] [Google Scholar]
- (44).Kells PM, Ouellet H, Santos-Aberturas J, Aparicio JF, Podust LM. Chem. Biol. 2010;17:841. doi: 10.1016/j.chembiol.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Tsukada S, Anzai Y, Li S, Kinoshita K, Sherman DH, Kato F. FEMS Microbiol. Lett. 2010;304:148. doi: 10.1111/j.1574-6968.2010.01899.x. [DOI] [PubMed] [Google Scholar]
- (46).Li S, Tietz DR, Rutaganira FU, Kells PM, Anzai Y, Kato F, Pochapsky TC, Sherman DH, Podust LM. J Biol Chem. 2012;287:37880. doi: 10.1074/jbc.M112.410340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wei JY, Pochapsky TC, Pochapsky SS. J. Am. Chem. Soc. 2005;127:6974. doi: 10.1021/ja051195j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Tripathi S, Li H, Poulos TL. Science. 2013;240:1227. doi: 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]
- (49).Fishelovitch D, Shaik S, Wolfson HJ, Nussinov R. J. Phys. Chem. B. 2010;114:5964. doi: 10.1021/jp101894k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.