Abstract
Aurora kinase A is a frequently amplified and overexpressed gene in in upper gastrointestinal adenocarcinomas (UGCs). Using in vitro cell models of UGCs, we investigated whether AURKA can regulate Signal Transducer and Activator of Transcription 3 (STAT3). Our data indicate that overexpression of AURKA in FLO-1 and AGS cells increase STAT3 phosphorylation at the Tyr705 site, whereas AURKA genetic depletion by siRNA results in decreased phosphorylation levels of STAT3 in FLO-1 and MKN45 cells. Immunofluorescence analysis showed that AURKA overexpression enhanced STAT3 nuclear translocation while AURKA genetic knockdown reduced the nuclear translocation of STAT3 in AGS and FLO-1 cells, respectively. Using a luciferase reporter assay, we demonstrated that AURKA expression induces transcriptional activity of STAT3. Pharmacological inhibition of AURKA by MLN8237 reduced STAT3 phosphorylation along with down-regulation of STAT3 pro-survival targets, BCL2 and MCL1. Moreover, by using clonogenic cells survival assay, we showed that MLN8237 single dose treatment reduced the ability of FLO-1 and AGS cells to form colonies. Additional experiments utilizing cell models of overexpression and knockdown of AURKA indicated that STAT3 upstream non-receptor tyrosine kinase Janus kinase 2 (JAK2) is mediating the effect of AURKA on STAT3. The inhibition of JAK2 using JAK2-specific inhibitor AZD1480 or siRNA knockdown, in presence of AURKA overexpression, abrogated the AURKA-mediated STAT3 activation. These results confirm that AURKA-JAK2 axis is the main mechanism by which AURKA regulates STAT3 activity. In conclusion, we report, for the first time, that AURKA promotes STAT3 activity through regulating the expression and phosphorylation levels of JAK2. This highlights the importance of targeting AURKA as a therapeutic approach to treat gastric and esophageal cancers.
1. Introduction
Upper gastrointestinal adenocarcinomas (UGCs), adenocarcinomas of the stomach and esophagus, are characterized by poor response to current chemotherapeutics (Hohenberger and Gretschel, 2003; Kelsen et al., 1998). Gastric cancer is the fourth most common malignancy and the second leading cause of cancer-related deaths in the world (Ferlay et al., 2010; Inoue and Tsugane, 2005), whereas esophageal cancer is the eighth most common cancer worldwide (Ferlay et al., 2010) and its incidence is continuously increasing (Devesa et al., 1998). Despite the recent advancement in understanding the biology of UGC, full elucidation of mechanisms and signaling molecules that promote tumorigenesis and survival remains elusive.
Aurora kinase A (AURKA) amplification and overexpression are frequent findings in UGCs as well as several other malignancies (Chung et al., 2005; Dar et al., 2008b; Marumoto et al., 2005; Sehdev et al., 2012). AURKA is a serine/threonine kinase that localizes to spindle poles and ensures its correct assembly in normal cells (Lens et al., 2010). Several studies have shown that AURKA overexpression promotes drug resistance and tumor recurrence (Cammareri et al., 2010; Otto et al., 2009; Yang et al., 2006). We and others reported that AURKA overexpression counteracts the functions of tumor suppressor genes p53 and p73 (Dar et al., 2008a; Katayama et al., 2012; Sehdev et al., 2014). In cancer cells, AURKA overexpression results in activating several oncogenic pathways including PI3K/AKT, β-catenin, and NF-κB (Dar et al., 2009; Katsha et al., 2013). These reports suggest that AURKA could serve as a signaling hub that connects and regulates several oncogenic signaling networks. The pharmacological inhibition of AURKA using small molecule inhibitor MLN8237, also known as alisertib, has shown significant inhibition of tumor growth in pre-clinical xenograft tumor models (Katsha et al., 2013; Sehdev et al., 2013) with promising results in phase II clinical trials (Friedberg et al., 2014; Matulonis et al., 2012).
Signal Transducer and Activator of Transcription 3 (STAT3) is an important transcription factor that regulates the expression of several cytokines, growth factors, and pro-survival genes that modulate various cellular events; including survival, cell cycle, invasion, and angiogenesis (Aggarwal et al., 2009; Bromberg, 2002; Kortylewski and Yu, 2007; Yu et al., 2007). STAT3 is activated through receptor tyrosine kinases and cytokines; and upon its activation, it dimerizes and translocates into the nucleus where it regulates its target genes (Jarnicki et al., 2010). In addition, non-receptor tyrosine kinases such as Janus kinase 2 (JAK2), can phosphorylate and activate STAT3 (Jarnicki et al., 2010; Kortylewski and Yu, 2007). Of note, STAT3 activation has been linked to chemoresistance in a number of malignancies including lung cancer (Kulesza et al., 2013), head and neck cancer (Bourguignon et al., 2012), and breast cancer (Lieblein et al., 2008). In gastric cancer, STAT3 activation has been associated with Helicobacter pylori infection and pre-neoplastic progression (Jackson et al., 2007). Furthermore, STAT3 activation is a common criterion of inflammatory and non-inflammatory gastric cancer mouse models (Giraud et al., 2012), which reflects the overall important role of STAT3 in gastric carcinogenesis.
In this report, we show that AURKA promotes STAT3 activity, nuclear translocation, and expression of pro-survival proteins (BCL2 and MCL1), possibly through regulating the JAK2-STAT3 axis. AURKA knockdown or pharmacological inhibition reversed these pro-survival effects. Collectively, our novel data demonstrate the importance of developing and moving AURKA specific inhibitors in clinical studies of upper gastrointestinal carcinomas.
2. Materials and Methods
2.1. Cell culture and reagents
Human gastric (AGS and MKN45) and esophageal (FLO-1) adenocarcinoma cell lines were maintained in Dulbecco’s modified Eagle’s medium (GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen Life Technologies, Carlsbad, CA) and 1% penicillin/streptomycin (GIBCO). AURKA investigational inhibitor MLN8237 (Millennium Pharmaceuticals, Inc., Cambridge, MA) was prepared and stored according to the manufacturer’s instructions. Specific antibodies against p-AURKA (Thr288), AURKA, p-STAT3 (Tyr705), STAT3, p-JAK2 (Tyr221), JAK2, BCL2, MCL1, and β-actin were purchased from Cell Signaling Technology (Beverly, MA). STAT3 inhibitor AZD1480 was purchased from Sigma-Aldrich (Milwaukee, WI). MLN8237 (alisertib) was purchased from Selleck Chemicals (Houston, TX). Transfection reagent Fugene 6 was purchased from Promega (Madison, WI).
2.2. AURKA expression and plasmids
Flag-tagged coding sequence of AURKA was sub-cloned into Xba I and BamH I sites of the adenoviral shuttle vector (pACCMV), and the recombinant adenovirus was generated by co-transfecting HEK-293AD cells with the shuttle and adenoviral backbone (pJM17) plasmids using the Calcium Phosphate Transfection Kit (Applied Biological Materials, Inc., Richmond, BC). The expression plasmid for AURKA was generated by PCR amplification of the full-length coding sequence of AURKA and cloned in frame into pcDNA3.1 (Invitrogen Life Technologies). A synthetic Flag-tag sequence was added at the N-terminus of AURKA. Cloning of AURKA was confirmed by sequencing and restriction enzyme digestion.
2.3. Western blotting
Cells were scraped, centrifuged, and pellets were re-suspended in lysis buffer (PBS and 1% Triton ×100) containing Halt Protease Inhibitor Cocktail and Halt Phosphatase Inhibitor Cocktail (Pierce Biotechnology, Inc., Rockford, IL) on ice. Protein concentration was measured using the Bradford Protein Assay (Bio-Rad Laboratories, Hercules, CA). Proteins (25 µg) from each sample were subjected to SDS/PAGE and transferred onto nitrocellulose membranes. Membranes were probed with specific primary antibodies and HRP-coupled secondary antibodies (Cell Signaling). Protein bands were visualized using a commercial Immobilon Western Chemiluminescent HRP Substrate kit (Millipore, Billerica, MA).
2.4. Immunofluorescence
AGS and FLO-1 cells were seeded in 8-well chambers (BD Falcon, Bedford, MA) and incubated overnight. Next day, AGS cells were infected with control or AURKA adenoviruses particles (5 MOI), whereas FLO-1 cells were transfected with scramble or AURKA siRNA (Cell Signaling) and cultured for 48h. Cells were washed with PBS and fixed with fresh 4% paraformaldehyde solution for 45 min at room temperature. After washing with ice-cold PBS, cells were incubated with permeabilization solution (0.1% Triton ×-100 in 0.1% Sodium Citrate) for 2 min on ice. Then, cells were washed again with PBS twice for 1 min each followed by incubation with 10% non-immune goat serum blocking solution (Invitrogen) for 20 min at room temperature. Next, cells were incubated in a specific primary antibody against STAT3 or AURKA diluted in PBS (1:400) for 2h at room temperature. After washing 3 times with ice-cold PBS, cells were incubated in fluorescein isothiocyanate (FITC)-tagged secondary antibody (1:1,000; Invitrogen) for 45 min at room temperature. Cells were then washed in PBS, mounted with Vectashield/DAPI (Vector Laboratories, Burlingame, CA), visualized, and randomly selected images were taken using an Olympus BX51 fluorescence microscope (Olympus Co., Japan). The percentage of nuclear STAT3-positive cells was calculated from at least 200 cells from each experiment using ImageJ software (http://www.uhnresearch.ca/facilities/wcif/imagej/).
2.5. Luciferase reporter assay
STAT-Luc reporter vector (Clontech, Mountain View, CA) was used to measure the activity of STAT3. Cells were seeded in 24-well plates and transiently co-transfected with 0.5 µg of STAT3-Luc reporter in combination with 0.5, 1.0 or 1.5 µg of AURKA vector or 1.0 µg of pcDNA3.1 vector using Fugene 6 according to the manufacturer’s instructions. In a separate experiment, MKN45 cells were transfected with STAT3-Luc reporter (0.5 µg) in combination with shAURKA (0.1, 0.25 or 0.5 µg) or shScramble (0.5 µg). Luciferase activity was measured using the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer’s instructions. For the rescue experiment, we transfected the FLO-1 cells with siScramble or siAURKA along with 0.5 µg STAT3-luc reporter and cells allowed to settle for 24h. Then, we transfected cells with 2.0 µg of pcDNA3.1 or AURKA-pcDNA3.1 vectors using Fugene 6. The STAT3 luciferase reporter activity was measured after an additional 24 h.
2.6. AURKA silencing by Small interfering RNA (siRNA)
Cells were transfected with siScramble, siAURKA or siJAK2 (Cell Signaling) for 24h in 0% FBS. Next day, 20% FBS-DMEM medium was added, and cells were allowed to recover for another 24h before harvesting. In a separate experiment, and in order to check whether AURKA reconstitution, after its knockdown, would restore STAT3 phosphorylation, we transfected the FLO-1 cells with siScramble or siAURKA for 24h. Then, we transfected cells with 2.0 µg of pcDNA3.1 or AURKA-pcDNA3.1 vectors using Fugene 6. Cells were harvested after an additional 24h and subjected to Western blot analysis.
2.7. Clonogenic cell survival assay
FLO-1 and AGS cells were seeded at 5000 cells/well in 6-well plates. Next day, cells were treated with 0.5 µM of MLN8237 for 24h. After washing with PBS, the cells were incubated in drug-free complete DMEM medium for 10 days. Subsequently, cell colonies were fixed with 2% Paraformaldehyde solution for 10 min, and stained overnight with crystal violet (0.05% Crystal Violet in 50% Methanol). Next, colonies were gently washed with PBS and photographed. Cell survival was determined by quantifying the dye signal in each well with ImageJ analysis software.
2.8. RNA extraction and real-time RT-PCR
Total RNA from cells was extracted using a miRNeasy extraction kit (Qiagen, Valencia, CA). RNA (2 µg) was converted to cDNA using an iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR (qRT-PCR) was performed using an iCycler (Bio-Rad) with the threshold cycle number determined by use of iCycler software version 2.1. Reactions were performed in triplicate, and the threshold cycle numbers were averaged. The results of the genes were normalized to HPRT1 housekeeping gene for both cells as described previously (El-Rifai et al., 2002). The primers used in qRT-PCR analysis are shown in Supplemental Table 1.
2.9. Statistical analysis
Data are presented as means ± standard error of mean. All in vitro experiments were carried out in triplicate. The statistical significance of the studies was determined by the Student’s t test using GraphPad Prism 5 software (GraphPad software, Inc. La Jolla, CA). Differences with P values ≤0.05 are considered significant.
3. Results
3.1. AURKA induces STAT3 phosphorylation and activity
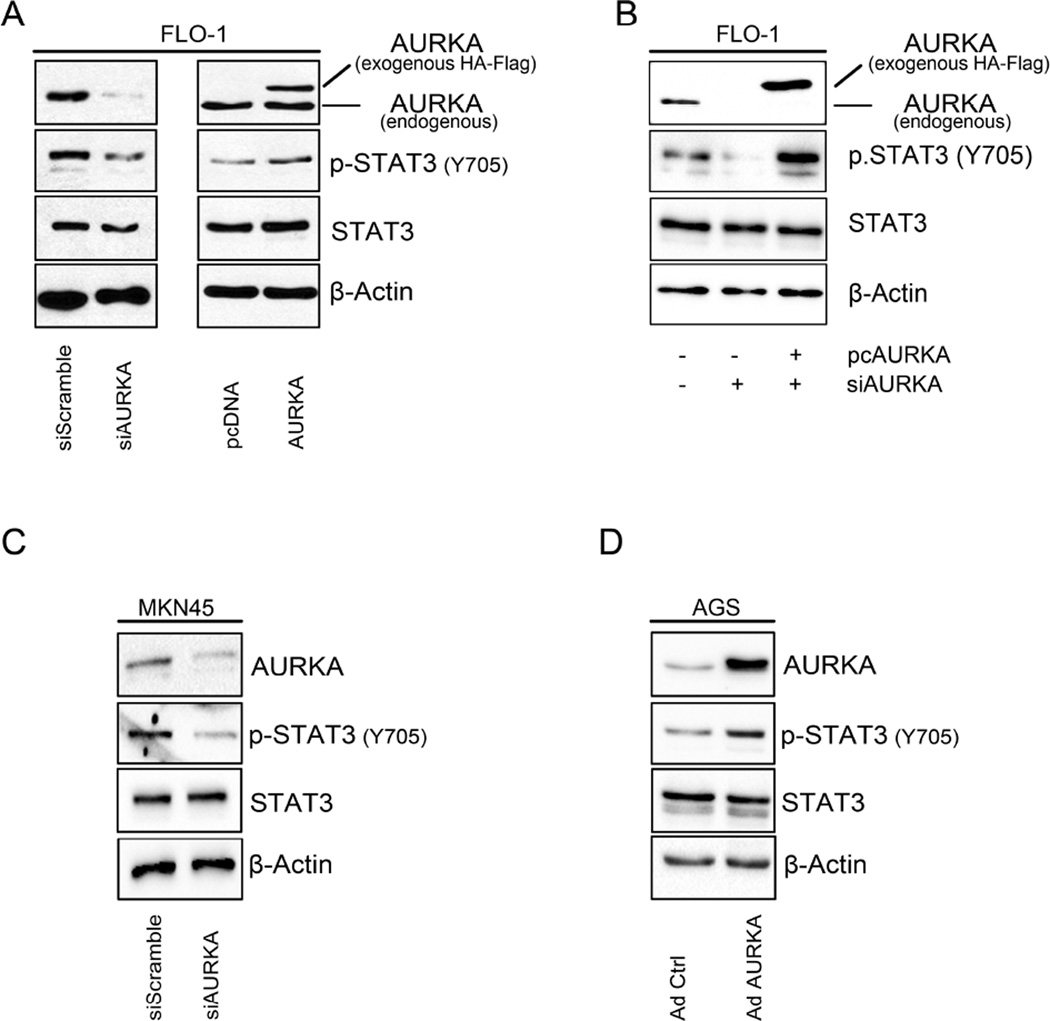
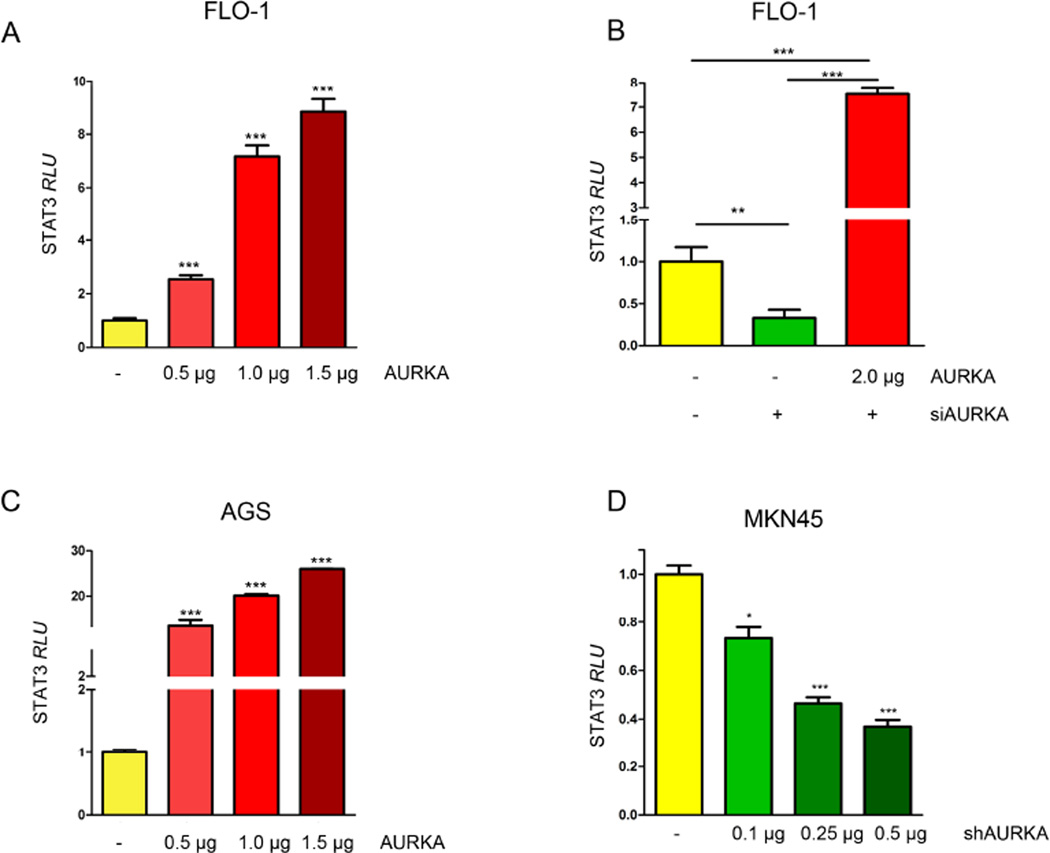
In order to investigate whether changes in AURKA levels would affect phosphorylation of STAT3, we modulated AURKA levels in FLO-1, AGS, and MKN45 cell lines and assessed the changes in STAT3 phosphorylation by Western blotting. Our data indicated that in FLO-1 cells, and after knocking down AURKA, p-STAT3 (Tyr705) levels were decreased (Figure 1A, left panel). On the other hand, FLO-1 cells stably overexpressing AURKA showed higher STAT3 phosphorylation compared to control cells (Figure 1A, right panel). In a rescue experiment, we confirmed that knockdown of endogenous AURKA by siRNA followed by overexpression of exogenous AURKA, restored AURKA-induced phosphorylation of STAT3 in FLO-1 cells (Figure 1B). To confirm these observations, we aslo knocked down endogenous AURKA in MKN45 cells and examined STAT3 phosphorylation. Indeed, Western blot analysis data showed a decrease in p-STAT3 levels (Tyr705), as compared to scramble control transfected cells (Figure 1C). Additionally, by using an adenovirus system we transiently overexpressed AURKA in AGS cells and evaluated p-STAT3 (Tyr705) levels. Our data indicated an increase in STAT3 phosphorylation in Adeno-AURKA infected cells as compared to control adenovirus infected cells (Figure 1D). Next, to investigate if the increase of STAT3 phosphorylation is accompanied with increased STAT3 activity, we transfected FLO-1 cells with 0.5, 1.0 or 1.5 µg of AURKA vector in combination with STAT3 luciferase reporter plasmid (0.5 µg), and STAT3 transcriptional activity was measured. Our data showed a dose-dependent significant induction of STAT3 activity (Figure 2A). For a rescue experiment, we knocked down the endogenous AURKA for 24h followed by its reconstitution using exogenous AURKA transfection. The STAT3 transcriptional activity was measured and demonstrated that knockdown of endogenous AURKA reduced the STAT3 reporter activity whereas overexpression of AURKA restored the activity of the STAT3 reporter (Figure 2B). In AGS cells, STAT3 activity showed dose-dependent increase in response to AURKA transfection (Figure 2C). Conversely, knockdown of endogenous AURKA in MKN45 cells reduced STAT3 transcriptional activity in a dose-dependent manner (Figure 2D). Together, these data suggest that AURKA promotes phosphorylation and activation of STAT3.
Figure 1. AURKA promotes phosphorylation of STAT3 in upper gastrointestinal cancer cells.
(A) AURKA knockdown (left panel) or stable overexpression (right panel) in FLO-1 cells led to a decrease or increase in p-STAT3 (Tyr705), respectively. (B) Knockdown of endogenous AURKA led to a decrease in the p-STAT3 (Tyr705). Reconstitution of AURKA, after knockdown of endogenous levels, restored the STAT3 phosphorylation levels. (C) AURKA knockdown in MKN45 cells resulted in decreased phosphorylation of STAT3 (Tyr705). (D) AGS cells showed increased p-STAT3 (Tyr705) protein level in response to transient AURKA overexpression.
Figure 2. AURKA overexpression induces STAT3 transcriptional activity.
(A) FLO-1 cells were transfected with pcDNA or AURKA in combination with STAT3 luciferase reporter plasmids, and STAT3 luciferase reporter activity was measured. The data indicate that AURKA can enhance STAT3 transcriptional activity. (B) Knockdown of endogenous AURKA decreases STAT3 luciferase reporter activity, whereas re-introducing AURKA rescued the STAT3 activity levels. (C) AGS cells were transfected with pcDNA or AURKA in combination with STAT3 luciferase reporter plasmids, and STAT3 luciferase reporter activity was measured. The data indicate that AURKA can enhance STAT3 transcriptional activity. (D) MKN45 cells were transfected with shScramble or shAURKA in combination with STAT3 luciferase reporter plasmids, and STAT3 luciferase reporter activity was assessed. The data show that knockdown of endogenous AURKA decreases STAT3 transcriptional activity in a dose-dependent fashion. and AGS
3.2. STAT3 nuclear translocation is affiliated with AURKA levels
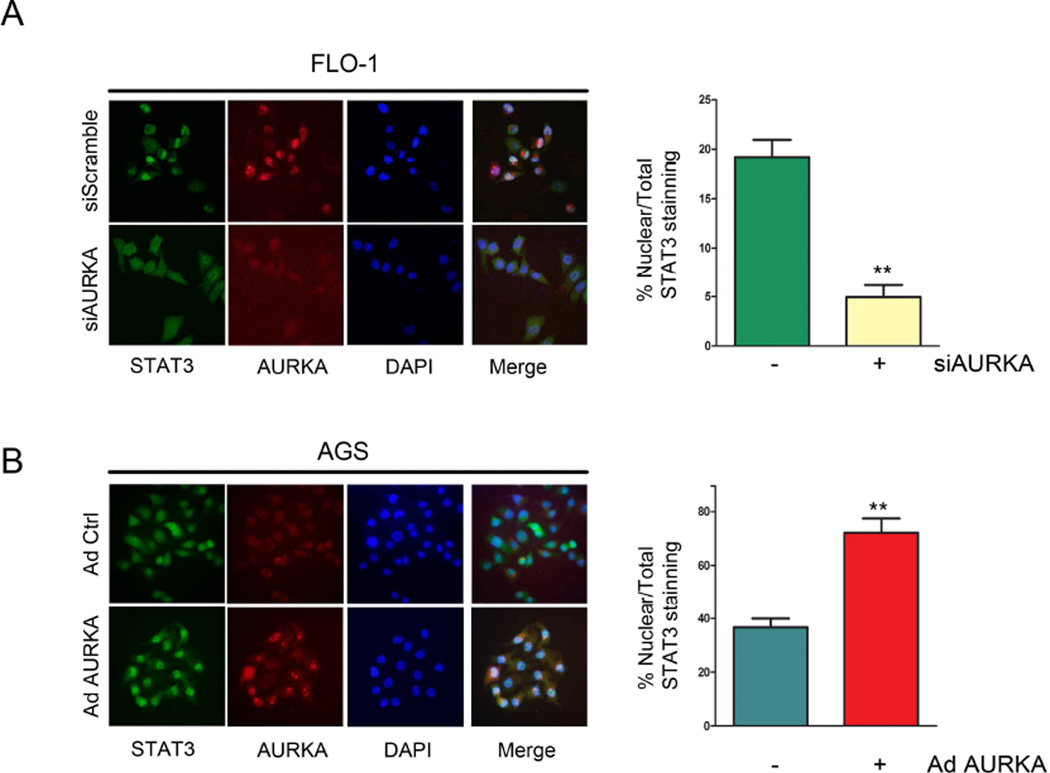
Upon phosphorylation, STAT3 translocates to the nucleus where it binds to and activates expression of its transcription target genes. To elucidate whether AURKA can alter STAT3 nuclear translocation, we performed immunofluorescence analysis for STAT3 in FLO-1 and AGS cells after AURKA knockdown or overexpression, respectively. Our data clearly indicated that AURKA knockdown in FLO-1 cells led to a significant (p<0.01) decrease of cells showing positive nuclear staining of STAT3 relative to control cells (Figure 3A). In contrast, AURKA overexpression in AGS cells resulted in a dramatic increase (p<0.01) of cells depicting positive nuclear staining of STAT3, as compared to control cells (Figure 3B). Collectively, these data clearly show that AURKA regulates phosphorylation and nuclear localization of STAT3.
Figure 3. AURKA expression modulates STAT3 nuclear translocation.
Representative images of STAT3 immunofluorescence analysis in FLO-1 cells (A) after AURKA knockdown, or in AGS cells (B) after AURKA overexpression by adenovirus are shown. Original magnification is shown at ×400. AURKA knockdown or overexpression significantly decreased (p<0.01) or increased (p<0.01) nuclear STAT3 positive staining relative to vehicle-treated control, respectively.
3.3. Pharmacological inhibition of AURKA attenuates STAT3 phosphorylation and suppresses cell survival
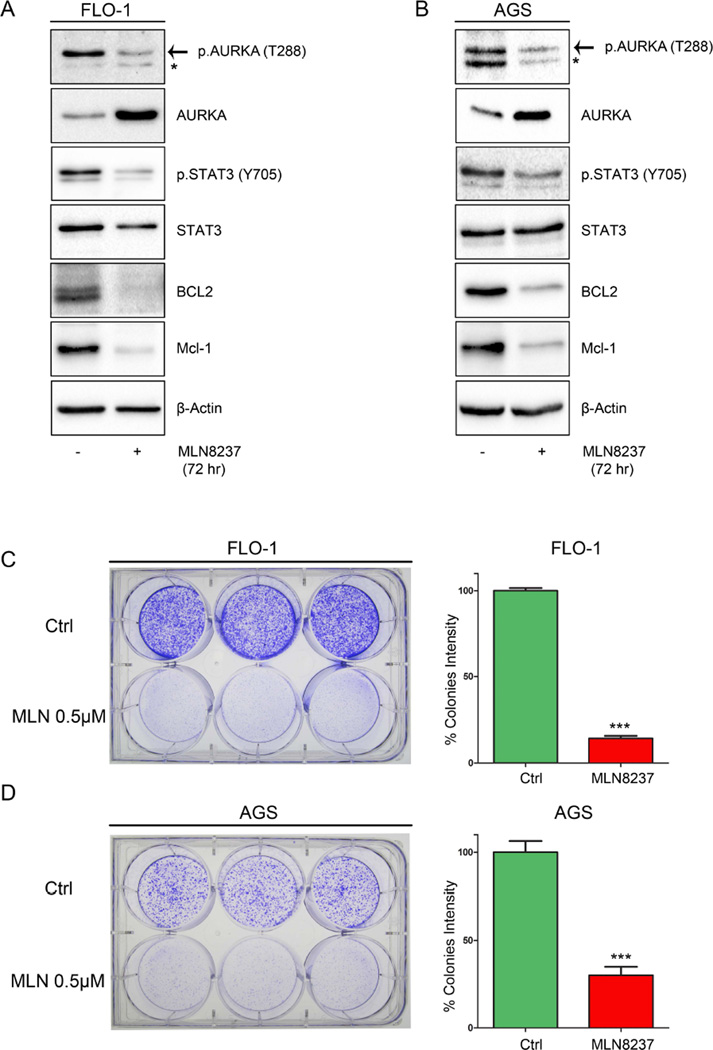
Based on the aforementioned data, we decided to examine whether AURKA kinase activity is involved in regulating STAT3. Therefore, we treated FLO-1, AGS and MKN45 cells with AURKA specific inhibitor MLN8237 (0.5 µM) for 30 min or 1h, and subjected the cell lysates to Western blot analysis. Our data showed that AURKA inhibition decreased STAT3 phosphorylation at Tyr705 in all cell lines (Supplemental Figure 1 A-C). Next, we treated FLO-1 and AGS cells for 72h with MLN8237 (0.5 µM) and examined the expression levels of pro-survival proteins, BCL2 and MCL1, known protein targets of STAT3 with anti-apoptotic functions (Bhattacharya et al., 2005; Liu et al., 2003). Indeed, our data indicated that MLN8237 treatment led to a decrease in STAT3 phosphorylation and BCL2 and MCL1 expression levels (Figure 4 A&B). The detected transient increase of total unphosphorylated AURKA protein level following MLN8237 treatment could be a cellular response to inhibition of its activity and function, which has been reported previously by others (Do et al., 2013; Li and Rana, 2012). To demonstrate the biological effect of AURKA inhibition on cell growth, we performed long-term clonogenic cell survival assay. AGS and FLO-1 cells were treated with MLN8237 (0.5 µM) for only 24h and cultured for 10 days. Our data showed that MLN8237 significantly impaired the cells ability to form colonies as indicated by the quantitative analysis at the end of the experiment (Figure 4 C&D). Collectively, these data suggest that AURKA inhibition suppresses the STAT3 pathway leading to decreased survival of cancer cells.
Figure 4. AURKA pharmacological inhibition decreases STAT3 phosphorylation and reduces cell survival.
FLO-1 (A) and AGS (B) cells treated with MLN8237 (0.5 µM) for 72h and subjected to Western blot analysis. Data indicated down-regulation of p-STAT3 (Tyr705) and STAT3 targets, BCL2 and MCL1, protein expression. FLO-1 (C) and AGS (D) cells treated with vehicle or MLN8237 (0.5 µM) for 24h and then cultured in fresh media for 10 days. Graphs show colonies quantitative data (right panels).
3.4. AURKA regulates phosphorylation and expression of JAK2
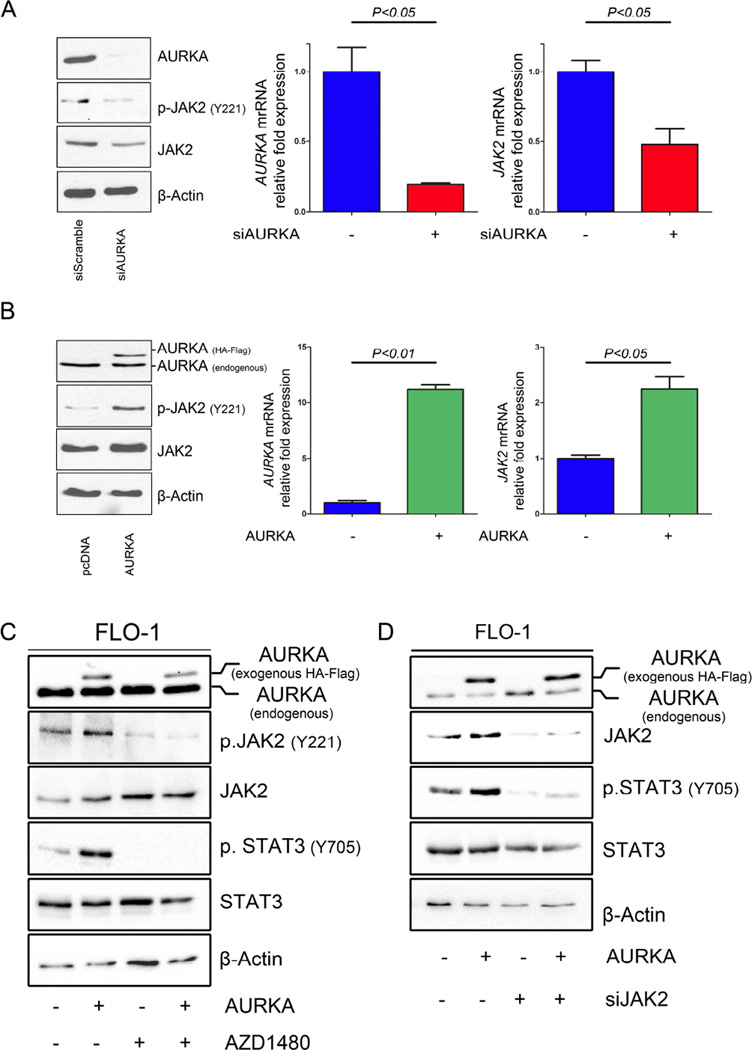
To elucidate the exact mechanism by which AURKA regulates STAT3, we examined STAT3 upstream molecules, specifically Janus kinase 2 (JAK2). Our results indicated for the first time that knockdown of AURKA by siRNA resulted in a significant decrease in phosphorylation and expression of JAK2 in FLO-1 (Figure 5A, left panel) and MKN45 (Supplemental Figure 2). To determine whether the decrease in JAK2 protein levels is initiated transcriptionally, we assessed JAK2 mRNA expression after knockdown or overexpression of AURKA by qRT-PCR in FLO-1 cells. Our data showed that AURKA knockdown resulted in decreased JAK2 mRNA levels (Figure 5A, right panel). Conversely, AURKA overexpression led to an increase in JAK2 phosphorylation at Y221 and JAK2 protein and mRNA levels (Figure 5B). Previously, it was reported that JAK2 specific inhibitor AZD1480 could block STAT3 phosphorylation and signaling (Hedvat et al., 2009; Scuto et al., 2011). Therefore, to determine if JAK2 is necessary for AURKA-mediated phosphorylation of STAT3, we treated FLO-1 cells overexpressing AURKA with AZD1480. Western blot analysis indicated that inhibition of JAK2 abrogated AURKA-induced phosphorylation of STAT3 (Figure 5C). Consistent with these results, knockdown of JAK2 in FLO-1 cells stably overexpressing AURKA significantly reduced the AURKA-induced STAT3 phosphorylation (Figure 5D). Taken together, these results clearly indicate that JAK2 mediates the effect of AURKA on STAT3.
Figure 5. AURKA regulates JAK2 expression.
(A) Silencing AURKA (siAURKA) in FLO-1 cells led to decreased JAK2 at both protein and mRNA levels. (B) FLO-1 cells stably overexpressing AURKA showed an increase in JAK2 at both protein and mRNA levels. (C-D) Inhibition of JAK2 using pharmacologic inhibitor (AZD1480) (C) or genetic knockdown JAK2 using siRNA (siJAK2) (D) abrogated AURKA-induced phosphorylation of STAT3 in FLO-1 cells.
4. Discussion
In this study, we demonstrate for the first time that AURKA regulates the phosphorylation, nuclear localization, and transcription activity of STAT3. Our results indicate that AURKA regulation of STAT3 is mediated by JAK2. Importantly, our data suggest that pharmacological inhibition of AURKA could be a plausible approach to suppress the AURKA-STAT3 oncogenic axis.
STAT3 plays a major role in tumorigenesis by mediating activation of several pro-oncogenic functions such as proliferation, survival, invasion, and angiogenesis (Yu et al., 2007; Yu et al., 2009). Several studies have shown that H. Pylori infection leads to STAT3 activation through regulation of IL-6 and IL-11 (Bronte-Tinkew et al., 2009; Lee et al., 2012). In esophageal cancer, it has been shown that STAT3 contributes to survival and proliferation of cancer cells (Wang et al., 2013; Zhang et al., 2012). These reports show the important role of STAT3 in gastric and esophageal cancer progression and thus, it is of high importance to target STAT3 as a therapeutic approach for both malignancies. To date, there are no active clinical trials to test the efficacy of STAT3 inhibitors in UGC (clinicaltrials.gov). In our current report, we show that AURKA knockdown or pharmacological inhibition can significantly decrease STAT3 phosphorylation/activity and expression of STAT3 downstream targets, BCL2 and MCL1. We also demonstrated that MLN8237 can suppress survival of AGS and FLO-1 cells, and it is possible that other physiological functions could be affected as well. Constitutive activation of STAT3 or activating mutations in its upstream molecule, JAK2, trigger tumorigenesis, thereby promoting invasive growth and metastasis (Yu et al., 2007). In our study, we showed that the levels and phosphorylation JAK2 were affected by AURKA expression, suggesting that AURKA regulates the JAK2-STAT3 signaling. On the other hand, a previous report showed that activating mutations in JAK2, up-regulate AURKA expression through c-Myc, and this promotes chemoresistance to cisplatin (Sumi et al., 2011). Taken together with our data, this may suggest the presence of a positive feedback loop through which the cancer cells maintain constant activation of STAT3 and overexpression of AURKA, collectively promoting cancer cell survival. In addition, AURKA can regulate cell migration and adhesion in ovarian cancer through regulation of SRC (Do et al., 2013), which is an upstream regulator of STAT3. This highlights the role of AURKA in modulating oncogenic signaling pathways related to STAT3 in cancer.
Our recent study has indicated that AURKA can also regulate the pro-inflammatory NF-κB signaling in gastric cancer (Katsha et al., 2013). STAT3 is known to play important roles in promoting the pro-inflammatory signaling (Yu et al., 2007; Yu et al., 2009). Therefore, it is possible that AURKA overexpression could be a major molecular event that promotes inflammation and carcinogenesis by activating NF-κB and STAT3 signaling pathways. Our current data clearly indicate that pharmacologic or genetic inhibition of AURKA abrogates the JAK2-STAT3 signaling axis. This finding is a significant addition to our knowledge about the role of AURKA in carcinogenesis that has been shown to implicate the regulation of other key signaling pathways such as β-catenin (Dar et al., 2009), NF-κB (Katsha et al., 2013), and p53 family proteins (Katayama et al., 2012). Collectively, these data suggest that AURKA is placed at a major signaling hub in carcinogenesis. Therefore, targeted inhibition of AURKA, which is capable of regulating several important pathways in cancer cells, could be a novel therapeutic approach to target multiple oncogenic signaling pathways. Of note, phase II clinical trials using MLN8237, also known as alisertib, demonstrated promising results (Friedberg et al., 2014; Matulonis et al., 2012). The implementation of this strategy could be successful in overcoming drug resistance and eliminating cancer cells in UGCs. However, a single agent therapy may not be successful and a combination with existing chemotherapy regimens may be more beneficial and need to be examined in clinical trials.
In summary, our results demonstrate the JAK2-STAT3 axis as a novel signaling pathway regulated by AURKA in upper gastrointestinal cancers. Ongoing early phase clinical trials and future studies are likely to provide important information for moving AURKA inhibitors into the clinical practice.
Supplementary Material
Highlights.
The relationship between AURKA and STAT3 in cancers of the stomach and esophagus was examined using in vitro models.
AURKA expression regulates STAT3 phosphorylation and activity.
JAK2 mediates AURKA induced phosphorylation of STAT3.
AURKA pharmacological inhibition down-regulates p.STAT3 and its target proteins.
Acknowledgments
Grant Support: This study was supported by R01CA131225 (WER) from the National Institute of Health; Vanderbilt SPORE in Gastrointestinal Cancer (P50 CA95103), Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or Vanderbilt University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contribution:
Ahmed Katsha: Design of experiments and acquisition of data; analysis and interpretation of data; drafting of the manuscript; technical and material support.
Janet Arras: Experimental support and acquisition of data
Mohammed Soutto: analysis and interpretation of data
Abbes Belkhiri: analysis and interpretation of data; experimental troubleshooting; drafting of the manuscript; critical revision of the manuscript
Wael El-Rifai: study concept and design; obtained funding; study supervision; experimental troubleshooting; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content
Disclosure of Potential Conflicts of Interest: All authors indicated “no conflict of interest”.
References
- Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, Dey S, Sung B. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Annals of the New York Academy of Sciences. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Ray RM, Johnson LR. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. The Biochemical journal. 2005;392:335–344. doi: 10.1042/BJ20050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J. Stat proteins and oncogenesis. The Journal of clinical investigation. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte-Tinkew DM, Terebiznik M, Franco A, Ang M, Ahn D, Mimuro H, Sasakawa C, Ropeleski MJ, Peek RM, Jr, Jones NL. Helicobacter pylori cytotoxin-associated gene A activates the signal transducer and activator of transcription 3 pathway in vitro and in vivo. Cancer research. 2009;69:632–639. doi: 10.1158/0008-5472.CAN-08-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammareri P, Scopelliti A, Todaro M, Eterno V, Francescangeli F, Moyer MP, Agrusa A, Dieli F, Zeuner A, Stassi G. Aurora-a is essential for the tumorigenic capacity and chemoresistance of colorectal cancer stem cells. Cancer research. 2010;70:4655–4665. doi: 10.1158/0008-5472.CAN-09-3953. [DOI] [PubMed] [Google Scholar]
- Chung CM, Man C, Jin Y, Jin C, Guan XY, Wang Q, Wan TS, Cheung AL, Tsao SW. Amplification and overexpression of aurora kinase A (AURKA) in immortalized human ovarian epithelial (HOSE) cells. Molecular carcinogenesis. 2005;43:165–174. doi: 10.1002/mc.20098. [DOI] [PubMed] [Google Scholar]
- Dar AA, Belkhiri A, Ecsedy J, Zaika A, El-Rifai W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer research. 2008a;68:8998–9004. doi: 10.1158/0008-5472.CAN-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AA, Belkhiri A, El-Rifai W. The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene. 2009;28:866–875. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AA, Zaika A, Piazuelo MB, Correa P, Koyama T, Belkhiri A, Washington K, Castells A, Pera M, El-Rifai W. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer. 2008b;112:1688–1698. doi: 10.1002/cncr.23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–2053. [PubMed] [Google Scholar]
- Do TV, Xiao F, Bickel LE, Klein-Szanto AJ, Pathak HB, Hua X, Howe C, O'Brien SW, Maglaty M, Ecsedy JA, Litwin S, Golemis EA, Schilder RJ, Godwin AK, Connolly DC. Aurora kinase A mediates epithelial ovarian cancer cell migration and adhesion. Oncogene. 2013 doi: 10.1038/onc.2012.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Rifai W, Moskaluk CA, Abdrabbo MK, Harper J, Yoshida C, Riggins GJ, Frierson HF, Jr, Powell SM. Gastric cancers overexpress S100A calcium-binding proteins. Cancer research. 2002;62:6823–6826. [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer. Journal international du cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Friedberg JW, Mahadevan D, Cebula E, Persky D, Lossos I, Agarwal AB, Jung J, Burack R, Zhou X, Leonard EJ, Fingert H, Danaee H, Bernstein SH. Phase II study of alisertib, a selective Aurora A kinase inhibitor, in relapsed and refractory aggressive B- and T-cell non-Hodgkin lymphomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:44–50. doi: 10.1200/JCO.2012.46.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud AS, Menheniott TR, Judd LM. Targeting STAT3 in gastric cancer. Expert opinion on therapeutic targets. 2012;16:889–901. doi: 10.1517/14728222.2012.709238. [DOI] [PubMed] [Google Scholar]
- Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, Armstrong B, Bebernitz G, Weng S, Wang L, Ye M, McEachern K, Chen H, Morosini D, Bell K, Alimzhanov M, Ioannidis S, McCoon P, Cao ZA, Yu H, Jove R, Zinda M. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–315. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- Inoue M, Tsugane S. Epidemiology of gastric cancer in Japan. Postgraduate medical journal. 2005;81:419–424. doi: 10.1136/pgmj.2004.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CB, Judd LM, Menheniott TR, Kronborg I, Dow C, Yeomans ND, Boussioutas A, Robb L, Giraud AS. Augmented gp130-mediated cytokine signalling accompanies human gastric cancer progression. The Journal of pathology. 2007;213:140–151. doi: 10.1002/path.2218. [DOI] [PubMed] [Google Scholar]
- Jarnicki A, Putoczki T, Ernst M. Stat3: linking inflammation to epithelial cancer - more than a "gut" feeling? Cell division. 2010;5:14. doi: 10.1186/1747-1028-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Wang J, Treekitkarnmongkol W, Kawai H, Sasai K, Zhang H, Wang H, Adams HP, Jiang S, Chakraborty SN, Suzuki F, Arlinghaus RB, Liu J, Mobley JA, Grizzle WE, Wang H, Sen S. Aurora kinase-A inactivates DNA damage-induced apoptosis and spindle assembly checkpoint response functions of p73. Cancer cell. 2012;21:196–211. doi: 10.1016/j.ccr.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsha A, Soutto M, Sehdev V, Peng D, Washington MK, Piazuelo MB, Tantawy MN, Manning HC, Lu P, Shyr Y, Ecsedy J, Belkhiri A, El-Rifai W. Aurora Kinase A Promotes Inflammation and Tumorigenesis in Mice and Human Gastric Neoplasia. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsen DP, Ginsberg R, Pajak TF, Sheahan DG, Gunderson L, Mortimer J, Estes N, Haller DG, Ajani J, Kocha W, Minsky BD, Roth JA. Chemotherapy followed by surgery compared with surgery alone for localized esophageal cancer. The New England journal of medicine. 1998;339:1979–1984. doi: 10.1056/NEJM199812313392704. [DOI] [PubMed] [Google Scholar]
- Kortylewski M, Yu H. Stat3 as a potential target for cancer immunotherapy. Journal of immunotherapy. 2007;30:131–139. doi: 10.1097/01.cji.0000211327.76266.65. [DOI] [PubMed] [Google Scholar]
- Kulesza DW, Carre T, Chouaib S, Kaminska B. Silencing of the transcription factor STAT3 sensitizes lung cancer cells to DNA damaging drugs, but not to TNFalpha- and NK cytotoxicity. Experimental cell research. 2013;319:506–516. doi: 10.1016/j.yexcr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Lee KS, Kalantzis A, Jackson CB, O'Connor L, Murata-Kamiya N, Hatakeyama M, Judd LM, Giraud AS, Menheniott TR. Helicobacter pylori CagA triggers expression of the bactericidal lectin REG3gamma via gastric STAT3 activation. PloS one. 2012;7:e30786. doi: 10.1371/journal.pone.0030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Li Z, Rana TM. A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nature communications. 2012;3:1085. doi: 10.1038/ncomms2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieblein JC, Ball S, Hutzen B, Sasser AK, Lin HJ, Huang TH, Hall BM, Lin J. STAT3 can be activated through paracrine signaling in breast epithelial cells. BMC cancer. 2008;8:302. doi: 10.1186/1471-2407-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ma Y, Cole SM, Zander C, Chen KH, Karras J, Pope RM. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- Matulonis UA, Sharma S, Ghamande S, Gordon MS, Del Prete SA, Ray-Coquard I, Kutarska E, Liu H, Fingert H, Zhou X, Danaee H, Schilder RJ. Phase II study of MLN8237 (alisertib), an investigational Aurora A kinase inhibitor, in patients with platinum-resistant or -refractory epithelial ovarian, fallopian tube, or primary peritoneal carcinoma. Gynecol Oncol. 2012;127:63–69. doi: 10.1016/j.ygyno.2012.06.040. [DOI] [PubMed] [Google Scholar]
- Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, Kenney AM, Schulte JH, Beijersbergen R, Christiansen H, Berwanger B, Eilers M. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Scuto A, Krejci P, Popplewell L, Wu J, Wang Y, Kujawski M, Kowolik C, Xin H, Chen L, Wang Y, Kretzner L, Yu H, Wilcox WR, Yen Y, Forman S, Jove R. The novel JAK inhibitor AZD1480 blocks STAT3 and FGFR3 signaling, resulting in suppression of human myeloma cell growth and survival. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2011;25:538–550. doi: 10.1038/leu.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehdev V, Katsha A, Arras J, Peng D, Soutto M, Ecsedy J, Zaika A, Belkhiri A, El-Rifai W. HDM2 regulation by AURKA promotes cell survival in gastric cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:76–86. doi: 10.1158/1078-0432.CCR-13-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehdev V, Katsha A, Ecsedy J, Zaika A, Belkhiri A, El-Rifai W. The combination of alisertib, an investigational Aurora kinase A inhibitor, and docetaxel promotes cell death and reduces tumor growth in preclinical cell models of upper gastrointestinal adenocarcinomas. Cancer. 2013;119:904–914. doi: 10.1002/cncr.27801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehdev V, Peng D, Soutto M, Washington MK, Revetta F, Ecsedy J, Zaika A, Rau TT, Schneider-Stock R, Belkhiri A, El-Rifai W. The aurora kinase A inhibitor MLN8237 enhances cisplatin-induced cell death in esophageal adenocarcinoma cells. Molecular cancer therapeutics. 2012;11:763–774. doi: 10.1158/1535-7163.MCT-11-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi K, Tago K, Kasahara T, Funakoshi-Tago M. Aurora kinase A critically contributes to the resistance to anti-cancer drug cisplatin in JAK2 V617F mutant-induced transformed cells. FEBS letters. 2011;585:1884–1890. doi: 10.1016/j.febslet.2011.04.068. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu S, Shen M, Liu J, Wang M, Li C, Wang Y, Deng A, Mei Q. STAT3 is involved in esophageal carcinogenesis through regulation of Oct-1. Carcinogenesis. 2013;34:678–688. doi: 10.1093/carcin/bgs361. [DOI] [PubMed] [Google Scholar]
- Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. International journal of cancer. Journal international du cancer. 2006;119:2304–2312. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nature reviews. Immunology. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du XL, Wang CJ, Lin DC, Ruan X, Feng YB, Huo YQ, Peng H, Cui JL, Zhang TT, Wang YQ, Zhang H, Zhan QM, Wang MR. Reciprocal activation between PLK1 and Stat3 contributes to survival and proliferation of esophageal cancer cells. Gastroenterology. 2012;142:521–530. doi: 10.1053/j.gastro.2011.11.023. e523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.