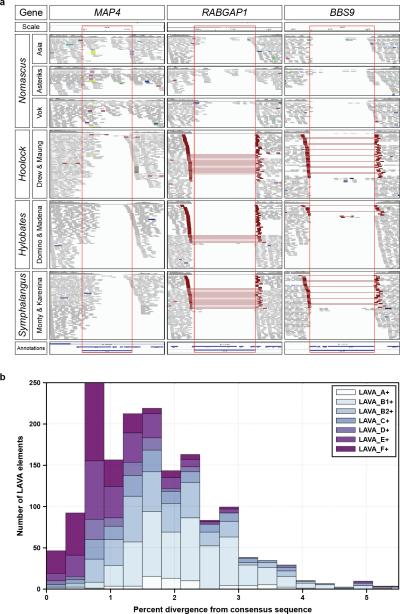
Extended Data Figure 4. Evolution of the LAVA element.
a, Screenshots from the Integrative Genomics Viewer (IGV) browser for loci MAP4, RABGAP1, and BBS9. Each column shows portions of the IGV visualization of a LAVA insertion locus identified in Nleu1.0 and its flanking sequence. Red rectangles indicate the margins of each LAVA insertion. Read pairs are colored in red when their insert size is larger-than-expected, indicating the presence of a LAVA insertion. MAP4 is a shared LAVA insertion, while RABGAP1 and BBS9 are Nomascus-specific. b, LAVA elements containing at least 300 bp of the LA section of LAVA elements were selected and reanalyzed using RepeatMasker to determine subfamily affiliation and divergence from the consensus sequence. LAVA elements are grouped based upon their subfamily affiliations (see legend top right). The x-axis shows the percent divergence from the respective consensus sequence, and the y-axis shows the number of elements with a certain percent divergence from the consensus sequence.